Note
Go to the end to download the full example code.
Sine bell and squared Sine bell window multiplication
In this example, we use sine bell or squared sine bell window multiplication to apodize a NMR signal in the time domain.
import spectrochempy as scp
DATADIR = scp.preferences.datadir
path = DATADIR / "nmrdata" / "bruker" / "tests" / "nmr" / "topspin_1d"
dataset1D = scp.read_topspin(path, expno=1, remove_digital_filter=True)
dataset1D
Normalize the dataset values and reduce the time domain
dataset1D /= dataset1D.real.data.max() # normalize
dataset1D = dataset1D[0.0:15000.0]
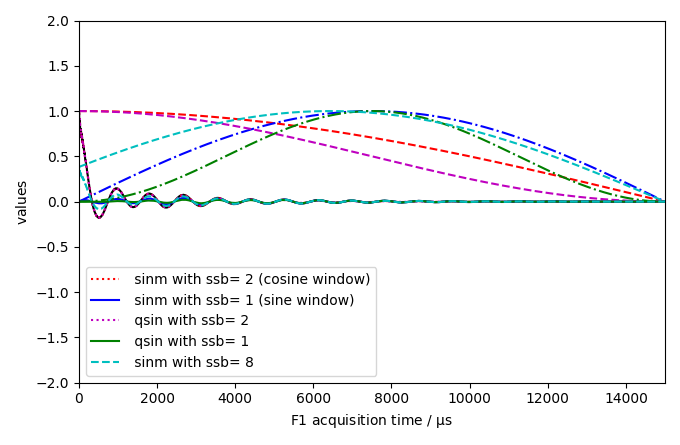
Apply Sine bell window apodization with parameter ssb=2, which correspond to a cosine function
this is equivalent to
or also
Apply Sine bell window apodization with parameter ssb=2, which correspond to a sine function
Apply Squared Sine bell window apodization with parameter ssb=1 and ssb=2
Apply shifted Sine bell window apodization with parameter ssb=8 (mixed sine/cosine window)
Plotting
dataset1D.plot(zlim=(-2, 2), color="k")
curve1.plot(color="r", clear=False)
new1.plot(
data_only=True, color="r", clear=False, label=" sinm with ssb= 2 (cosine window)"
)
curve2.plot(color="b", clear=False)
new2.plot(
data_only=True, color="b", clear=False, label=" sinm with ssb= 1 (sine window)"
)
curve3.plot(color="m", clear=False)
new3.plot(data_only=True, color="m", clear=False, label=" qsin with ssb= 2")
curve4.plot(color="g", clear=False)
new4.plot(data_only=True, color="g", clear=False, label=" qsin with ssb= 1")
curve5.plot(color="c", ls="--", clear=False)
new5.plot(
data_only=True,
color="c",
ls="--",
clear=False,
label=" sinm with ssb= 8",
legend="best",
)
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python scp.show() sphinx_gallery_thumbnail_number = -1
Total running time of the script: (0 minutes 0.403 seconds)