spectrochempy.NDDataset
- class NDDataset(data=None, coordset=None, coordunits=None, coordtitles=None, **kwargs)[source]
The main N-dimensional dataset class used by
SpectroChemPy.The
NDDatasetis the main object use by SpectroChemPy. Like numpyndarray’s,NDDatasethave the capability to be sliced, sorted and subject to mathematical operations. But, in addition,NDDatasetmay have units, can be masked and each dimensions can have coordinates also with units. This makeNDDatasetaware of unit compatibility, e.g., for binary operation such as additions or subtraction or during the application of mathematical operations. In addition or in replacement of numerical data for coordinates,NDDatasetcan also have labeled coordinates where labels can be different kind of objects (str,datetime,ndarrayor otherNDDataset’s, etc…).- Parameters:
data (array-like) – Data array contained in the object. The data can be a list, a tuple, a
ndarray, a subclass ofndarray, anotherNDDatasetor aCoordobject. Any size or shape of data is accepted. If not given, an emptyNDDatasetwill be inited. At the initialisation the provided data will be eventually cast to andarray. If the provided objects is passed which already contains some mask, or units, these elements will be used if possible to accordingly set those of the created object. If possible, the provided data will not be copied fordatainput, but will be passed by reference, so you should make a copy of thedatabefore passing them if that’s the desired behavior or set thecopyargument toTrue.coordset (
CoordSetinstance, optional) – It contains the coordinates for the different dimensions of thedata. ifCoordSetis provided, it must specify thecoordandlabelsfor all dimensions of thedata. Multiplecoord’s can be specified in aCoordSetinstance for each dimension.coordunits (
list, optional, default:None) – A list of units corresponding to the dimensions in the order of the coordset.coordtitles (
list, optional, default:None) – A list of titles corresponding of the dimensions in the order of the coordset.**kwargs – Optional keyword parameters (see Other Parameters).
- Other Parameters:
dtype (
strordtype, optional, default:np.float64) – If specified, the data will be cast to this dtype, else the data will be cast to float64 or complex128.dims (
listofstr, optional) – If specified the list must have a length equal to the number od data dimensions (ndim) and the elements must be taken amongx,y,z,u,v,w or t. If not specified, the dimension names are automatically attributed in this order.name (
str, optional) – A user-friendly name for this object. If not given, the automaticidgiven at the object creation will be used as a name.labels (array-like of objects, optional) – Labels for the
data. labels can be used only for 1D-datasets. The labels array may have an additional dimension, meaning several series of labels for the same data. The given array can be a list, a tuple, andarray, a ndarray-like, aNDArrayor any subclass ofNDArray.mask (array-like of
boolorNOMASK, optional) – Mask for the data. The mask array must have the same shape as the data. The given array can be a list, a tuple, or andarray. Each values in the array must beFalsewhere the data are valid and True when they are not (like in numpy masked arrays). Ifdatais already aMaskedArray, or any array object (such as aNDArrayor subclass of it), providing amaskhere, will cause the mask from the masked array to be ignored.units (
Unitinstance orstr, optional) – Units of the data. If data is aQuantitythenunitsis set to the unit of thedata; if a unit is also explicitly provided an error is raised. Handling of units use the pint package.timezone (
datetime.tzinfo, optional) – The timezone where the data were created. If not specified, the local timezone is assumed.title (
str, optional) – The title of the data dimension. Thetitleattribute should not be confused with thename. Thetitleattribute is used for instance for labelling plots of the data. It is optional but recommended to give a title to each ndarray data.meta (
dict-like object, optional) – Additional metadata for this object. Must be dict-like but no further restriction is placed on meta.author (
str, optional) – Name(s) of the author(s) of this dataset. By default, name of the computer note where this dataset is created.description (
str, optional) – An optional description of the nd-dataset. A shorter alias isdesc.origin (
str, optional) – Origin of the data: Name of organization, address, telephone number, name of individual contributor, etc., as appropriate.roi (
list) – Region of interest (ROI) limits.history (
str, optional) – A string to add to the object history.copy (
bool, optional) – Perform a copy of the passed object. Default is False.
Notes
The underlying array in a
NDDatasetobject can be accessed through thedataattribute, which will return a conventionalndarray.Attributes Summary
The array with imaginary-imaginary component of hypercomplex 2D data.
The array with imaginary-real component of hypercomplex 2D
data.The array with real-imaginary component of hypercomplex 2D
data.The array with real component in both dimension of hypercomplex 2D
data.Transposed
NDDataset.Acquisition date.
Creator of the dataset (str).
The main matplotlib axe associated to this dataset.
The matplotlib axe associated to the transposed dataset.
Matplotlib colorbar axe associated to this dataset.
Matplotlib colorbar axe associated to the transposed dataset.
Matplotlib projection x axe associated to this dataset.
Matplotlib projection y axe associated to this dataset.
Provides a comment (Alias to the description attribute).
List of the
Coordnames.CoordSetinstance.List of the
Coordtitles.List of the
Coordunits.Creation date object (Datetime).
The
dataarray.Provides a description of the underlying data (str).
True if the
dataarray is dimensionless - Readonly property (bool).Names of the dimensions (list).
Get current directory for this dataset.
Matplotlib plot divider.
Return the data type.
Matplotlib figure associated to this dataset.
Matplotlib figure associated to this dataset.
Get current filename for this dataset.
Type of current file.
True if at least one of the
dataarray dimension is complex.True if the
dataarray is not empty.True is the name has been defined (bool).
True if the
dataarray have units - Readonly property (bool).Describes the history of actions made on this array (List of strings).
Object identifier - Readonly property (str).
The array with imaginary component of the
data.True if the
dataarray has only one dimension (bool).True if the 'data' are complex (Readonly property).
True if the
dataarray is empty or size=0, and if no label are present.True if the
dataare real values - Readonly property (bool).True if the
dataare integer values - Readonly property (bool).True if the
dataarray is hypercomplex with interleaved data.True if the
dataarray have labels - Readonly property (bool).True if the
dataarray has masked values.True if the
dataarray is hypercomplex .Range of the data.
Return the local timezone.
Data array (
ndarray).Data array (
ndarray).Mask for the data (
ndarrayof bool).The actual masked
dataarray - Readonly property (numpy.ma.ndarray).Additional metadata (
Meta).ndarray- models data.Date of modification (readonly property).
A user-friendly name (str).
A dictionary containing all the axes of the current figures.
The number of dimensions of the
dataarray (Readonly property).Origin of the data.
Projectinstance.The array with real component of the
data.Region of interest (ROI) limits (list).
A tuple with the size of each dimension - Readonly property.
Size of the underlying
dataarray - Readonly property (int).Filename suffix.
Return the timezone information.
An user-friendly title (str).
The actual array with mask and unit (
Quantity).bool- True if thedatadoes not haveunits(Readonly property).Unit- The units of the data.Alias of
values.Quantity- The actual values (data, units) contained in this object (Readonly property).Methods Summary
abs(dataset[, dtype])Calculate the absolute value of the given NDDataset element-wise.
absolute(dataset[, dtype])Calculate the absolute value of the given NDDataset element-wise.
add_coordset(*coords[, dims])Add one or a set of coordinates from a dataset.
all(dataset[, dim, keepdims])Test whether all array elements along a given axis evaluate to True.
amax(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
amin(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
any(dataset[, dim, keepdims])Test whether any array element along a given axis evaluates to True.
arange([start, stop, step, dtype])Return evenly spaced values within a given interval.
argmax(dataset[, dim])Indexes of maximum of data along axis.
argmin(dataset[, dim])Indexes of minimum of data along axis.
around(dataset[, decimals])Evenly round to the given number of decimals.
Make data and mask (ndim >= 1) laid out in Fortran order in memory.
astype([dtype])Cast the data to a specified type.
atleast_2d([inplace])Expand the shape of an array to make it at least 2D.
average(dataset[, dim, weights, keepdims])Compute the weighted average along the specified axis.
clip(dataset[, a_min, a_max])Clip (limit) the values in a dataset.
Close a Matplotlib figure associated to this dataset.
component([select])Take selected components of an hypercomplex array (RRR, RIR, ...).
conj(dataset[, dim])Conjugate of the NDDataset in the specified dimension.
conjugate(dataset[, dim])Conjugate of the NDDataset in the specified dimension.
coord([dim])Return the coordinates along the given dimension.
coordmax(dataset[, dim])Find coordinates of the maximum of data along axis.
coordmin(dataset[, dim])Find oordinates of the mainimum of data along axis.
copy([deep, keepname])Make a disconnected copy of the current object.
cumsum(dataset[, dim, dtype])Return the cumulative sum of the elements along a given axis.
Delete all coordinate settings.
diag(dataset[, offset])Extract a diagonal or construct a diagonal array.
diagonal(dataset[, offset, dim, dtype])Return the diagonal of a 2D array.
dump(filename, **kwargs)Save the current object into compressed native spectrochempy format.
empty(shape[, dtype])Return a new
NDDatasetof given shape and type, without initializing entries.empty_like(dataset[, dtype])Return a new uninitialized
NDDataset.eye(N[, M, k, dtype])Return a 2-D array with ones on the diagonal and zeros elsewhere.
fromfunction(cls, function[, shape, dtype, ...])Construct a nddataset by executing a function over each coordinate.
fromiter(iterable[, dtype, count])Create a new 1-dimensional array from an iterable object.
full(shape[, fill_value, dtype])Return a new
NDDatasetof given shape and type, filled withfill_value.full_like(dataset[, fill_value, dtype])Return a
NDDatasetof fill_value.geomspace(start, stop[, num, endpoint, dtype])Return numbers spaced evenly on a log scale (a geometric progression).
get_axis(*args, **kwargs)Determine an axis index whatever the syntax used (axis index or dimension names).
get_labels([level])Get the labels at a given level.
identity(n[, dtype])Return the identity
NDDatasetof a given shape.is_units_compatible(other)Check the compatibility of units with another object.
ito(other[, force])Inplace scaling to different units.
Inplace rescaling to base units.
Quantity scaled in place to reduced units, inplace.
linspace(cls, start, stop[, num, endpoint, ...])Return evenly spaced numbers over a specified interval.
load(filename, **kwargs)Open data from a '.scp' (NDDataset) or '.pscp' (Project) file.
loads(js, Any])Deserialize dataset from JSON.
logspace(cls, start, stop[, num, endpoint, ...])Return numbers spaced evenly on a log scale.
max(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
mean(dataset[, dim, dtype, keepdims])Compute the arithmetic mean along the specified axis.
min(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
ones(shape[, dtype])Return a new
NDDatasetof given shape and type, filled with ones.ones_like(dataset[, dtype])Return
NDDatasetof ones.pipe(func, *args, **kwargs)Apply func(self, *args, **kwargs).
plot([method])Plot the dataset using the specified method.
ptp(dataset[, dim, keepdims])Range of values (maximum - minimum) along a dimension.
random([size, dtype])Return random floats in the half-open interval [0.0, 1.0).
Remove all masks previously set on this array.
round(dataset[, decimals])Evenly round to the given number of decimals.
round_(dataset[, decimals])Evenly round to the given number of decimals.
save(**kwargs)Save dataset in native .scp format.
save_as(filename, **kwargs)Save the current NDDataset in SpectroChemPy format (.scp).
set_complex([inplace])Set the object data as complex.
set_coordset(*args, **kwargs)Set one or more coordinates at once.
set_coordtitles(*args, **kwargs)Set titles of the one or more coordinates.
set_coordunits(*args, **kwargs)Set units of the one or more coordinates.
set_hypercomplex([inplace])Alias of set_quaternion.
set_quaternion([inplace])Alias of set_quaternion.
sort(**kwargs)Return the dataset sorted along a given dimension.
squeeze(*dims[, inplace])Remove single-dimensional entries from the shape of a NDDataset.
std(dataset[, dim, dtype, ddof, keepdims])Compute the standard deviation along the specified axis.
sum(dataset[, dim, dtype, keepdims])Sum of array elements over a given axis.
swapaxes(dim1, dim2[, inplace])Alias of
swapdims.swapdims(dim1, dim2[, inplace])Interchange two dimensions of a NDDataset.
take(indices, **kwargs)Take elements from an array.
to(other[, inplace, force])Return the object with data rescaled to different units.
to_array()Return a numpy masked array.
to_base_units([inplace])Return an array rescaled to base units.
to_reduced_units([inplace])Return an array scaled in place to reduced units.
Convert a NDDataset instance to an
DataArrayobject.transpose(*dims[, inplace])Permute the dimensions of a NDDataset.
var(dataset[, dim, dtype, ddof, keepdims])Compute the variance along the specified axis.
zeros(shape[, dtype])Return a new
NDDatasetof given shape and type, filled with zeros.zeros_like(dataset[, dtype])Return a
NDDatasetof zeros.Attributes Documentation
- II
The array with imaginary-imaginary component of hypercomplex 2D data.
(Readonly property).
- RR
The array with real component in both dimension of hypercomplex 2D
data.This readonly property is equivalent to the
realproperty.
- acquisition_date
Acquisition date.
- author
Creator of the dataset (str).
- ax
The main matplotlib axe associated to this dataset.
- axT
The matplotlib axe associated to the transposed dataset.
- axec
Matplotlib colorbar axe associated to this dataset.
- axecT
Matplotlib colorbar axe associated to the transposed dataset.
- axex
Matplotlib projection x axe associated to this dataset.
- axey
Matplotlib projection y axe associated to this dataset.
- comment
Provides a comment (Alias to the description attribute).
- coordset
CoordSetinstance.Contains the coordinates of the various dimensions of the dataset. It’s a readonly property. Use set_coords to change one or more coordinates at once.
- coordtitles
List of the
Coordtitles.Read only property. Use set_coordtitle to eventually set titles.
- coordunits
List of the
Coordunits.Read only property. Use set_coordunits to eventually set units.
- created
Creation date object (Datetime).
- description
Provides a description of the underlying data (str).
- dimensionless
True if the
dataarray is dimensionless - Readonly property (bool).Notes
Dimensionlessis different ofunitlesswhich means no unit.
- dims
Names of the dimensions (list).
The name of the dimensions are ‘x’, ‘y’, ‘z’…. depending on the number of dimension.
- directory
Get current directory for this dataset.
- divider
Matplotlib plot divider.
- dtype
Return the data type.
- fig
Matplotlib figure associated to this dataset.
- fignum
Matplotlib figure associated to this dataset.
- filename
Get current filename for this dataset.
- filetype
Type of current file.
- has_defined_name
True is the name has been defined (bool).
- has_units
True if the
dataarray have units - Readonly property (bool).See also
unitlessTrue if the data has no unit.
dimensionlessTrue if the data have dimensionless units.
- history
Describes the history of actions made on this array (List of strings).
- id
Object identifier - Readonly property (str).
- is_complex
True if the ‘data’ are complex (Readonly property).
- is_empty
True if the
dataarray is empty or size=0, and if no label are present.Readonly property (bool).
- labels
An array of labels for
data(ndarrayof str).An array of objects of any type (but most generally string), with the last dimension size equal to that of the dimension of data. Note that’s labelling is possible only for 1D data. One classical application is the labelling of coordinates to display informative strings instead of numerical values.
- limits
Range of the data.
- local_timezone
Return the local timezone.
- m
Data array (
ndarray).If there is no data but labels, then the labels are returned instead of data.
- magnitude
Data array (
ndarray).If there is no data but labels, then the labels are returned instead of data.
- meta
Additional metadata (
Meta).
- modified
Date of modification (readonly property).
- Returns:
str – Date of modification in isoformat.
- name
A user-friendly name (str).
When no name is provided, the
idof the object is returned instead.
- ndaxes
A dictionary containing all the axes of the current figures.
- origin
Origin of the data.
e.g. spectrometer or software
- roi
Region of interest (ROI) limits (list).
- shape
A tuple with the size of each dimension - Readonly property.
The number of
dataelement on each dimension (possibly complex). For only labelled array, there is no data, so it is the 1D and the size is the size of the array of labels.
- size
Size of the underlying
dataarray - Readonly property (int).The total number of data elements (possibly complex or hypercomplex in the array).
- suffix
Filename suffix.
Read Only property - automatically set when the filename is updated if it has a suffix, else give the default suffix for the given type of object.
- timezone
Return the timezone information.
A timezone’s offset refers to how many hours the timezone is from Coordinated Universal Time (UTC).
In spectrochempy, all datetimes are stored in UTC, so that conversion must be done during the display of these datetimes using tzinfo.
- title
An user-friendly title (str).
When the title is provided, it can be used for labeling the object, e.g., axe title in a matplotlib plot.
Methods Documentation
- abs(dataset, dtype=None)[source]
Calculate the absolute value of the given NDDataset element-wise.
absis a shorthand for this function. For complex input, a + ib, the absolute value is \(\sqrt{ a^2 + b^2}\) .- Parameters:
dataset (
NDDatasetor array-like) – Input array or object that can be converted to an array.dtype (dtype) – The type of the output array. If dtype is not given, infer the data type from the other input arguments.
- Returns:
NDDataset– The absolute value of each element in dataset.
- absolute(dataset, dtype=None)[source]
Calculate the absolute value of the given NDDataset element-wise.
absis a shorthand for this function. For complex input, a + ib, the absolute value is \(\sqrt{ a^2 + b^2}\) .- Parameters:
dataset (
NDDatasetor array-like) – Input array or object that can be converted to an array.dtype (dtype) – The type of the output array. If dtype is not given, infer the data type from the other input arguments.
- Returns:
NDDataset– The absolute value of each element in dataset.
- add_coordset(*coords, dims=None, **kwargs)[source]
Add one or a set of coordinates from a dataset.
- Parameters:
*coords (iterable) – Coordinates object(s).
dims (list) – Name of the coordinates.
**kwargs – Optional keyword parameters passed to the coordset.
- all(dataset, dim=None, keepdims=False)[source]
Test whether all array elements along a given axis evaluate to True.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or str, optional) – Axis or axes along which a logical AND reduction is performed. The default (
axis=None) is to perform a logical AND over all the dimensions of the input array.axismay be negative, in which case it counts from the last to the first axis.keepdims (bool, optional) – If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array. If the default value is passed, then
keepdimswill not be passed through to theallmethod of sub-classes ofndarray, however any non-default value will be. If the sub-class’ method does not implementkeepdimsany exceptions will be raised.
- Returns:
all – A new boolean or array is returned unless
outis specified, in which case a reference tooutis returned.
See also
anyTest whether any element along a given axis evaluates to True.
Notes
Not a Number (NaN), positive infinity and negative infinity evaluate to
Truebecause these are not equal to zero.
- amax(dataset, dim=None, keepdims=False, **kwargs)[source]
Return the maximum of the dataset or maxima along given dimensions.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or dimension name or tuple of int or dimensions, optional) – Dimension or dimensions along which to operate. By default, flattened input is used. If this is a tuple, the maximum is selected over multiple dimensions, instead of a single dimension or all the dimensions as before.
keepdims (bool, optional) – If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
amax – Maximum of the data. If
dimis None, the result is a scalar value. Ifdimis given, the result is an array of dimensionndim - 1.
See also
aminThe minimum value of a dataset along a given dimension, propagating NaNs.
minimumElement-wise minimum of two datasets, propagating any NaNs.
maximumElement-wise maximum of two datasets, propagating any NaNs.
fmaxElement-wise maximum of two datasets, ignoring any NaNs.
fminElement-wise minimum of two datasets, ignoring any NaNs.
argmaxReturn the indices or coordinates of the maximum values.
argminReturn the indices or coordinates of the minimum values.
Notes
For dataset with complex or hypercomplex type type, the default is the value with the maximum real part.
- amin(dataset, dim=None, keepdims=False, **kwargs)[source]
Return the maximum of the dataset or maxima along given dimensions.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or dimension name or tuple of int or dimensions, optional) – Dimension or dimensions along which to operate. By default, flattened input is used. If this is a tuple, the minimum is selected over multiple dimensions, instead of a single dimension or all the dimensions as before.
keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
amin – Minimum of the data. If
dimis None, the result is a scalar value. Ifdimis given, the result is an array of dimensionndim - 1.
See also
amaxThe maximum value of a dataset along a given dimension, propagating NaNs.
minimumElement-wise minimum of two datasets, propagating any NaNs.
maximumElement-wise maximum of two datasets, propagating any NaNs.
fmaxElement-wise maximum of two datasets, ignoring any NaNs.
fminElement-wise minimum of two datasets, ignoring any NaNs.
argmaxReturn the indices or coordinates of the maximum values.
argminReturn the indices or coordinates of the minimum values.
- any(dataset, dim=None, keepdims=False)[source]
Test whether any array element along a given axis evaluates to True.
Returns single boolean unless
dimis notNone- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or tuple of ints, optional) – Axis or axes along which a logical OR reduction is performed. The default (
axis=None) is to perform a logical OR over all the dimensions of the input array.axismay be negative, in which case it counts from the last to the first axis.keepdims (bool, optional) – If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array. If the default value is passed, then
keepdimswill not be passed through to theanymethod of sub-classes ofndarray, however any non-default value will be. If the sub-class’ method does not implementkeepdimsany exceptions will be raised.
- Returns:
any – A new boolean or
ndarrayis returned.
See also
allTest whether all array elements along a given axis evaluate to True.
- arange(start=0, stop=None, step=None, dtype=None, **kwargs)[source]
Return evenly spaced values within a given interval.
Values are generated within the half-open interval [start, stop).
- Parameters:
start (number, optional) – Start of interval. The interval includes this value. The default start value is 0.
stop (number) – End of interval. The interval does not include this value, except in some cases where step is not an integer and floating point round-off affects the length of out. It might be prefereble to use inspace in such case.
step (number, optional) – Spacing between values. For any output out, this is the distance between two adjacent values, out[i+1] - out[i]. The default step size is 1. If step is specified as a position argument, start must also be given.
dtype (dtype) – The type of the output array. If dtype is not given, infer the data type from the other input arguments.
**kwargs – Keywords argument used when creating the returned object, such as units, name, title, …
- Returns:
arange – Array of evenly spaced values.
See also
linspaceEvenly spaced numbers with careful handling of endpoints.
Examples
>>> scp.arange(1, 20.0001, 1, units='s', name='mycoord') NDDataset: [float64] s (size: 20)
- around(dataset, decimals=0)[source]
Evenly round to the given number of decimals.
- Parameters:
dataset (
NDDataset) – Input dataset.decimals (int, optional) – Number of decimal places to round to (default: 0). If decimals is negative, it specifies the number of positions to the left of the decimal point.
- Returns:
rounded_array – NDDataset containing the rounded values. The real and imaginary parts of complex numbers are rounded separately. The result of rounding a float is a float. If the dataset contains masked data, the mask remain unchanged.
See also
numpy.round,around,spectrochempy.round,spectrochempy.around,methods.,ceil,fix,floor,rint,trunc
- astype(dtype=None, **kwargs)[source]
Cast the data to a specified type.
- Parameters:
dtype (str or dtype) – Typecode or data-type to which the array is cast.
- atleast_2d(inplace=False)[source]
Expand the shape of an array to make it at least 2D.
- Parameters:
inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDDataset– The input array, but with dimensions increased.
See also
squeezeThe inverse operation, removing singleton dimensions.
- average(dataset, dim=None, weights=None, keepdims=False)[source]
Compute the weighted average along the specified axis.
- Parameters:
dataset (array_like) – Array containing data to be averaged.
dim (None or int or dimension name or tuple of int or dimensions, optional) – Dimension or dimensions along which to operate. By default, flattened input is used. If this is a tuple, the minimum is selected over multiple dimensions, instead of a single dimension or all the dimensions as before.
weights (array_like, optional) – An array of weights associated with the values in
dataset. Each value inacontributes to the average according to its associated weight. The weights array can either be 1-D (in which case its length must be the size ofdatasetalong the given axis) or of the same shape asdataset. Ifweights=None, then all data indatasetare assumed to have a weight equal to one. The 1-D calculation is:avg = sum(a * weights) / sum(weights)
The only constraint on
weightsis thatsum(weights)must not be 0.
- Returns:
average, – Return the average along the specified axis.
- Raises:
ZeroDivisionError – When all weights along axis are zero. See
numpy.ma.averagefor a version robust to this type of error.TypeError – When the length of 1D
weightsis not the same as the shape ofaalong axis.
See also
meanCompute the arithmetic mean along the specified axis.
Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.average(nd) <Quantity(1.25085858, 'absorbance')> >>> m = scp.average(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.x Coord: [float64] cm⁻¹ (size: 5549) >>> m = scp.average(nd, dim='y', weights=np.arange(55)) >>> m.data array([ 1.789, 1.789, ..., 1.222, 1.22])
- clip(dataset, a_min=None, a_max=None, **kwargs)[source]
Clip (limit) the values in a dataset.
Given an interval, values outside the interval are clipped to the interval edges. For example, if an interval of
[0, 1]is specified, values smaller than 0 become 0, and values larger than 1 become 1.No check is performed to ensure
a_min < a_max.- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
a_min (scalar or array_like or None) – Minimum value. If None, clipping is not performed on lower interval edge. Not more than one of
a_minanda_maxmay be None.a_max (scalar or array_like or None) – Maximum value. If None, clipping is not performed on upper interval edge. Not more than one of
a_minanda_maxmay be None. Ifa_minora_maxare array_like, then the three arrays will be broadcasted to match their shapes.
- Returns:
clip – An array with the elements of
a, but where values <a_minare replaced witha_min, and those >a_maxwitha_max.
- component(select='REAL')[source]
Take selected components of an hypercomplex array (RRR, RIR, …).
- Parameters:
select (str, optional, default=’REAL’) – If ‘REAL’, only real component in all dimensions will be selected. ELse a string must specify which real (R) or imaginary (I) component has to be selected along a specific dimension. For instance, a string such as ‘RRI’ for a 2D hypercomplex array indicated that we take the real component in each dimension except the last one, for which imaginary component is preferred.
- Returns:
component – Component of the complex or hypercomplex array.
- conj(dataset, dim='x')[source]
Conjugate of the NDDataset in the specified dimension.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (int, str, optional, default=(0,)) – Dimension names or indexes along which the method should be applied.
- Returns:
conjugated – Same object or a copy depending on the
inplaceflag.
See also
conj,real,imag,RR,RI,IR,II,part,set_complex,is_complex
- conjugate(dataset, dim='x')[source]
Conjugate of the NDDataset in the specified dimension.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (int, str, optional, default=(0,)) – Dimension names or indexes along which the method should be applied.
- Returns:
conjugated – Same object or a copy depending on the
inplaceflag.
See also
conj,real,imag,RR,RI,IR,II,part,set_complex,is_complex
- coord(dim='x')[source]
Return the coordinates along the given dimension.
- Parameters:
dim (int or str) – A dimension index or name, default index =
x. If an integer is provided, it is equivalent to theaxisparameter for numpy array.- Returns:
Coord– Coordinates along the given axis.
- copy(deep=True, keepname=False, **kwargs)[source]
Make a disconnected copy of the current object.
- Parameters:
deep (bool, optional) – If True a deepcopy is performed which is the default behavior.
keepname (bool) – If True keep the same name for the copied object.
- Returns:
object – An exact copy of the current object.
Examples
>>> nd1 = scp.NDArray([1. + 2.j, 2. + 3.j]) >>> nd1 NDArray: [complex128] unitless (size: 2) >>> nd2 = nd1 >>> nd2 is nd1 True >>> nd3 = nd1.copy() >>> nd3 is not nd1 True
- cumsum(dataset, dim=None, dtype=None)[source]
Return the cumulative sum of the elements along a given axis.
- Parameters:
dataset (array_like) – Calculate the cumulative sum of these values.
dim (None or int or dimension name , optional) – Dimension or dimensions along which to operate. By default, flattened input is used.
dtype (dtype, optional) – Type to use in computing the standard deviation. For arrays of integer type the default is float64, for arrays of float types it is the same as the array type.
- Returns:
sum – A new array containing the cumulative sum.
See also
Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.sum(nd) <Quantity(381755.783, 'absorbance')> >>> scp.sum(nd, keepdims=True) NDDataset: [float64] a.u. (shape: (y:1, x:1)) >>> m = scp.sum(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.data array([ 100.7, 100.7, ..., 74, 73.98])
- diag(dataset, offset=0, **kwargs)[source]
Extract a diagonal or construct a diagonal array.
See the more detailed documentation for
numpy.diagonalif you use this function to extract a diagonal and wish to write to the resulting array; whether it returns a copy or a view depends on what version of numpy you are using.- Parameters:
dataset (array_like) – If
datasetis a 2-D array, return a copy of itsk-th diagonal. Ifdatasetis a 1-D array, return a 2-D array withvon thek-th. diagonal.offset (int, optional) – Diagonal in question. The default is 0. Use offset>0 for diagonals above the main diagonal, and offset<0 for diagonals below the main diagonal.
- Returns:
diag – The extracted diagonal or constructed diagonal array.
- diagonal(dataset, offset=0, dim='x', dtype=None, **kwargs)[source]
Return the diagonal of a 2D array.
As we reduce a 2D to a 1D we must specified which is the dimension for the coordinates to keep!.
- Parameters:
dataset (
NDDatasetor array-like) – Object from which to extract the diagonal.offset (int, optional) – Offset of the diagonal from the main diagonal. Can be positive or negative. Defaults to main diagonal (0).
dim (str, optional) – Dimension to keep for coordinates. By default it is the last (-1,
xor another name if the default dimension name has been modified).dtype (dtype, optional) – The type of the returned array.
**kwargs – Additional keyword parameters to be passed to the NDDataset constructor.
- Returns:
diagonal – The diagonal of the input array.
See also
diagExtract a diagonal or construct a diagonal array.
Examples
>>> nd = scp.full((2, 2), 0.5, units='s', title='initial') >>> nd NDDataset: [float64] s (shape: (y:2, x:2)) >>> nd.diagonal(title='diag') NDDataset: [float64] s (size: 2)
- dump(filename, **kwargs)[source]
Save the current object into compressed native spectrochempy format.
- Parameters:
filename (str of
pathlibobject) – File name where to save the current object.
- empty(shape, dtype=None, **kwargs)[source]
Return a new
NDDatasetof given shape and type, without initializing entries.- Parameters:
shape (int or tuple of int) – Shape of the empty array.
dtype (data-type, optional) – Desired output data-type.
**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
empty – Array of uninitialized (arbitrary) data of the given shape, dtype, and order. Object arrays will be initialized to None.
- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
zeros_likeReturn an array of zeros with shape and type of input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
full_likeFill an array with shape and type of input.
zerosReturn a new array setting values to zero.
onesReturn a new array setting values to 1.
fullFill a new array.
Notes
empty, unlikezeros, does not set the array values to zero, and may therefore be marginally faster. On the other hand, it requires the user to manually set all the values in the array, and should be used with caution.Examples
>>> scp.empty([2, 2], dtype=int, units='s') NDDataset: [int64] s (shape: (y:2, x:2))
- empty_like(dataset, dtype=None, **kwargs)[source]
Return a new uninitialized
NDDataset.The returned
NDDatasethave the same shape and type as a given array. Units, coordset, … can be added in kwargs.- Parameters:
dataset (
NDDatasetor array-like) – Object from which to copy the array structure.dtype (data-type, optional) – Overrides the data type of the result.
**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
emptylike – Array of
fill_valuewith the same shape and type asdataset.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
Notes
This function does not initialize the returned array; to do that use for instance
zeros_like,ones_likeorfull_likeinstead. It may be marginally faster than the functions that do set the array values.
- eye(N, M=None, k=0, dtype=float, **kwargs)[source]
Return a 2-D array with ones on the diagonal and zeros elsewhere.
- Parameters:
N (int) – Number of rows in the output.
M (int, optional) – Number of columns in the output. If None, defaults to
N.k (int, optional) – Index of the diagonal: 0 (the default) refers to the main diagonal, a positive value refers to an upper diagonal, and a negative value to a lower diagonal.
dtype (data-type, optional) – Data-type of the returned array.
**kwargs – Other parameters to be passed to the object constructor (units, coordset, mask …).
- Returns:
eye – NDDataset of shape (N,M) An array where all elements are equal to zero, except for the
k-th diagonal, whose values are equal to one.
See also
Examples
>>> scp.eye(2, dtype=int) NDDataset: [float64] unitless (shape: (y:2, x:2)) >>> scp.eye(3, k=1, units='km').values <Quantity([[ 0 1 0] [ 0 0 1] [ 0 0 0]], 'kilometer')>
- fromfunction(cls, function, shape=None, dtype=float, units=None, coordset=None, **kwargs)[source]
Construct a nddataset by executing a function over each coordinate.
The resulting array therefore has a value
fn(x, y, z)at coordinate(x, y, z).- Parameters:
function (callable) – The function is called with N parameters, where N is the rank of
shapeor from the providedCoordSet.shape ((N,) tuple of ints, optional) – Shape of the output array, which also determines the shape of the coordinate arrays passed to
function. It is optional only ifCoordSetis None.dtype (data-type, optional) – Data-type of the coordinate arrays passed to
function. By default,dtypeis float.units (str, optional) – Dataset units. When None, units will be determined from the function results.
coordset (
CoordSetinstance, optional) – If provided, this determine the shape and coordinates of each dimension of the returnedNDDataset. If shape is also passed it will be ignored.**kwargs – Other kwargs are passed to the final object constructor.
- Returns:
fromfunction – The result of the call to
functionis passed back directly. Therefore the shape offromfunctionis completely determined byfunction.
See also
fromiterMake a dataset from an iterable.
Examples
Create a 1D NDDataset from a function
>>> func1 = lambda t, v: v * t >>> time = scp.Coord.arange(0, 60, 10, units='min') >>> d = scp.fromfunction(func1, v=scp.Quantity(134, 'km/hour'), coordset=scp.CoordSet(t=time)) >>> d.dims ['t'] >>> d NDDataset: [float64] km (size: 6)
- fromiter(iterable, dtype=np.float64, count=-1, **kwargs)[source]
Create a new 1-dimensional array from an iterable object.
- Parameters:
iterable (iterable object) – An iterable object providing data for the array.
dtype (data-type) – The data-type of the returned array.
count (
int, optional) – The number of items to read from iterable. The default is -1, which means all data is read.**kwargs – Other kwargs are passed to the final object constructor.
- Returns:
fromiter – The output nddataset.
See also
fromfunctionConstruct a nddataset by executing a function over each coordinate.
Notes
- Specify count to improve performance. It allows fromiter to pre-allocate the
output array, instead of resizing it on demand.
Examples
>>> iterable = (x * x for x in range(5)) >>> d = scp.fromiter(iterable, float, units='km') >>> d NDDataset: [float64] km (size: 5) >>> d.data array([ 0, 1, 4, 9, 16])
- full(shape, fill_value=0.0, dtype=None, **kwargs)[source]
Return a new
NDDatasetof given shape and type, filled withfill_value.- Parameters:
shape (int or sequence of ints) – Shape of the new array, e.g.,
(2, 3)or2.fill_value (scalar) – Fill value.
dtype (data-type, optional) – The desired data-type for the array, e.g.,
np.int8. Default is fill_value.dtype.**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
full – Array of
fill_value.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
zeros_likeReturn an array of zeros with shape and type of input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
full_likeFill an array with shape and type of input.
zerosReturn a new array setting values to zero.
onesReturn a new array setting values to one.
emptyReturn a new uninitialized array.
Examples
>>> scp.full((2, ), np.inf) NDDataset: [float64] unitless (size: 2) >>> scp.NDDataset.full((2, 2), 10, dtype=np.int) NDDataset: [int64] unitless (shape: (y:2, x:2))
- full_like(dataset, fill_value=0.0, dtype=None, **kwargs)[source]
Return a
NDDatasetof fill_value.The returned
NDDatasethave the same shape and type as a given array. Units, coordset, … can be added in kwargs- Parameters:
dataset (
NDDatasetor array-like) – Object from which to copy the array structure.fill_value (scalar) – Fill value.
dtype (data-type, optional) – Overrides the data type of the result.
**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
fulllike – Array of
fill_valuewith the same shape and type asdataset.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
zeros_likeReturn an array of zeros with shape and type of input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
zerosReturn a new array setting values to zero.
onesReturn a new array setting values to one.
emptyReturn a new uninitialized array.
fullFill a new array.
Examples
3 possible ways to call this method
from the API
>>> x = np.arange(6, dtype=int) >>> scp.full_like(x, 1) NDDataset: [float64] unitless (size: 6)
as a classmethod
>>> x = np.arange(6, dtype=int) >>> scp.NDDataset.full_like(x, 1) NDDataset: [float64] unitless (size: 6)
as an instance method
>>> scp.NDDataset(x).full_like(1, units='km') NDDataset: [float64] km (size: 6)
- geomspace(start, stop, num=50, endpoint=True, dtype=None, **kwargs)[source]
Return numbers spaced evenly on a log scale (a geometric progression).
This is similar to
logspace, but with endpoints specified directly. Each output sample is a constant multiple of the previous.- Parameters:
start (number) – The starting value of the sequence.
stop (number) – The final value of the sequence, unless
endpointis False. In that case,num + 1values are spaced over the interval in log-space, of which all but the last (a sequence of lengthnum) are returned.num (int, optional) – Number of samples to generate. Default is 50.
endpoint (bool, optional) – If true,
stopis the last sample. Otherwise, it is not included. Default is True.dtype (dtype) – The type of the output array. If
dtypeis not given, infer the data type from the other input arguments.**kwargs – Keywords argument used when creating the returned object, such as units, name, title, …
- Returns:
geomspace –
numsamples, equally spaced on a log scale.
- get_axis(*args, **kwargs)[source]
Determine an axis index whatever the syntax used (axis index or dimension names).
- Parameters:
dim, axis, dims (str, int, or list of str or index) – The axis indexes or dimensions names - they can be specified as argument or using keyword ‘axis’, ‘dim’ or ‘dims’.
negative_axis (bool, optional, default=False) – If True a negative index is returned for the axis value (-1 for the last dimension, etc…).
allows_none (bool, optional, default=False) – If True, if input is none then None is returned.
only_first (bool, optional, default: True) – By default return only information on the first axis if dim is a list. Else, return a list for axis and dims information.
- Returns:
axis (int) – The axis indexes.
dim (str) – The axis name.
- get_labels(level=0)[source]
Get the labels at a given level.
Used to replace
datawhen only labels are provided, and/or for labeling axis in plots.- Parameters:
level (int, optional, default:0) – Label level.
- Returns:
ndarray– The labels at the desired level or None.
- identity(n, dtype=None, **kwargs)[source]
Return the identity
NDDatasetof a given shape.The identity array is a square array with ones on the main diagonal.
- Parameters:
n (int) – Number of rows (and columns) in
nxnoutput.dtype (data-type, optional) – Data-type of the output. Defaults to
float.**kwargs – Other parameters to be passed to the object constructor (units, coordset, mask …).
- Returns:
identity –
nxnarray with its main diagonal set to one, and all other elements 0.
See also
Examples
>>> scp.identity(3).data array([[ 1, 0, 0], [ 0, 1, 0], [ 0, 0, 1]])
- is_units_compatible(other)[source]
Check the compatibility of units with another object.
- Parameters:
other (
ndarray) – The ndarray object for which we want to compare units compatibility.- Returns:
result – True if units are compatible.
Examples
>>> nd1 = scp.NDDataset([1. + 2.j, 2. + 3.j], units='meters') >>> nd1 NDDataset: [complex128] m (size: 2) >>> nd2 = scp.NDDataset([1. + 2.j, 2. + 3.j], units='seconds') >>> nd1.is_units_compatible(nd2) False >>> nd1.ito('minutes', force=True) >>> nd1.is_units_compatible(nd2) True >>> nd2[0].values * 60. == nd1[0].values True
- ito(other, force=False)[source]
Inplace scaling to different units. (same as
towith inplace= True).- Parameters:
See also
toRescaling of the current object data to different units.
to_base_unitsRescaling of the current object data to different units.
ito_base_unitsInplace rescaling of the current object data to different units.
to_reduced_unitsRescaling to reduced units.
ito_reduced_unitsRescaling to reduced units.
- ito_base_units()[source]
Inplace rescaling to base units.
See also
toRescaling of the current object data to different units.
itoInplace rescaling of the current object data to different units.
to_base_unitsRescaling of the current object data to different units.
to_reduced_unitsRescaling to redunced units.
ito_reduced_unitsInplace rescaling to reduced units.
- ito_reduced_units()[source]
Quantity scaled in place to reduced units, inplace.
Scaling to reduced units means one unit per dimension. This will not reduce compound units (e.g., ‘J/kg’ will not be reduced to m**2/s**2).
See also
toRescaling of the current object data to different units.
itoInplace rescaling of the current object data to different units.
to_base_unitsRescaling of the current object data to different units.
ito_base_unitsInplace rescaling of the current object data to different units.
to_reduced_unitsRescaling to reduced units.
- linspace(cls, start, stop, num=50, endpoint=True, retstep=False, dtype=None, **kwargs)[source]
Return evenly spaced numbers over a specified interval.
Returns num evenly spaced samples, calculated over the interval [start, stop]. The endpoint of the interval can optionally be excluded.
- Parameters:
start (array_like) – The starting value of the sequence.
stop (array_like) – The end value of the sequence, unless endpoint is set to False. In that case, the sequence consists of all but the last of num + 1 evenly spaced samples, so that stop is excluded. Note that the step size changes when endpoint is False.
num (int, optional) – Number of samples to generate. Default is 50. Must be non-negative.
endpoint (bool, optional) – If True, stop is the last sample. Otherwise, it is not included. Default is True.
retstep (bool, optional) – If True, return (samples, step), where step is the spacing between samples.
dtype (dtype, optional) – The type of the array. If dtype is not given, infer the data type from the other input arguments.
**kwargs – Keywords argument used when creating the returned object, such as units,
name, title, …
- Returns:
linspace (ndarray) – There are num equally spaced samples in the closed interval [start, stop] or the half-open interval [start, stop) (depending on whether endpoint is True or False).
step (float, optional) – Only returned if retstep is True Size of spacing between samples.
- classmethod load(filename: str | pathlib.Path | BinaryIO, **kwargs: Any) -> Any: """ Open data from a '*.scp' (NDDataset) or '*.pscp' (Project) file. Parameters ---------- filename : `str`, `pathlib` or `file` objects The name of the file to read (or a file objects). **kwargs Optional keyword parameters (see Other Parameters). Other Parameters ---------------- content : str, optional The optional content of the file(s) to be loaded as a binary string. See Also -------- read : Import dataset from various orgines. save : Save the current dataset. Notes ----- Adapted from `numpy.load` . Examples -------- >>> nd1 = scp.read('irdata/nh4y-activation.spg') >>> f = nd1.save() >>> f.name 'nh4y-activation.scp' >>> nd2 = scp.load(f) Alternatively, this method can be called as a class method of NDDataset or Project object: >>> from spectrochempy import * >>> nd2 = NDDataset.load(f) """ content = kwargs.get("content") if content: fid = io.BytesIO(content) else: # be sure to convert filename to a pathlib object with the # default suffix filename = pathclean(filename) suffix = cls().suffix filename = filename.with_suffix(suffix) if kwargs.get("directory") is not None: filename = pathclean(kwargs.get("directory")) / filename if not filename.exists()[source]
Open data from a ‘.scp’ (NDDataset) or ‘.pscp’ (Project) file.
- Parameters:
- Other Parameters:
content (str, optional) – The optional content of the file(s) to be loaded as a binary string.
Notes
Adapted from
numpy.load.Examples
>>> nd1 = scp.read('irdata/nh4y-activation.spg') >>> f = nd1.save() >>> f.name 'nh4y-activation.scp' >>> nd2 = scp.load(f)
Alternatively, this method can be called as a class method of NDDataset or Project object:
>>> from spectrochempy import * >>> nd2 = NDDataset.load(f)
- classmethod loads(js: dict[str, Any]) -> Any: """ Deserialize dataset from JSON. Parameters ---------- js : dict[str, Any] JSON object to deserialize Returns ------- Any Deserialized dataset object Raises ------ TypeError If JSON cannot be properly deserialized """ from spectrochempy.core.dataset.coord import Coord from spectrochempy.core.dataset.coordset import CoordSet from spectrochempy.core.dataset.nddataset import NDDataset from spectrochempy.core.project.project import Project from spectrochempy.core.script import Script # ......................... def item_to_attr(obj: Any, dic: dict[str, Any]) -> Any: for key, val in dic.items()[source]
Deserialize dataset from JSON.
- Parameters:
js (dict[str, Any]) – JSON object to deserialize
- Returns:
Any – Deserialized dataset object
- Raises:
TypeError – If JSON cannot be properly deserialized
- logspace(cls, start, stop, num=50, endpoint=True, base=10.0, dtype=None, **kwargs)[source]
Return numbers spaced evenly on a log scale.
In linear space, the sequence starts at
base ** start(baseto the power ofstart) and ends withbase ** stop(seeendpointbelow).- Parameters:
start (array_like) –
base ** startis the starting value of the sequence.stop (array_like) –
base ** stopis the final value of the sequence, unlessendpointis False. In that case,num + 1values are spaced over the interval in log-space, of which all but the last (a sequence of lengthnum) are returned.num (int, optional) – Number of samples to generate. Default is 50.
endpoint (bool, optional) – If true,
stopis the last sample. Otherwise, it is not included. Default is True.base (float, optional) – The base of the log space. The step size between the elements in
ln(samples) / ln(base)(orlog_base(samples)) is uniform. Default is 10.0.dtype (dtype) – The type of the output array. If
dtypeis not given, infer the data type from the other input arguments.**kwargs – Keywords argument used when creating the returned object, such as units, name, title, …
- Returns:
logspace –
numsamples, equally spaced on a log scale.
See also
arangeSimilar to linspace, with the step size specified instead of the number of samples. Note that, when used with a float endpoint, the endpoint may or may not be included.
linspaceSimilar to logspace, but with the samples uniformly distributed in linear space, instead of log space.
geomspaceSimilar to logspace, but with endpoints specified directly.
- max(dataset, dim=None, keepdims=False, **kwargs)[source]
Return the maximum of the dataset or maxima along given dimensions.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or dimension name or tuple of int or dimensions, optional) – Dimension or dimensions along which to operate. By default, flattened input is used. If this is a tuple, the maximum is selected over multiple dimensions, instead of a single dimension or all the dimensions as before.
keepdims (bool, optional) – If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
amax – Maximum of the data. If
dimis None, the result is a scalar value. Ifdimis given, the result is an array of dimensionndim - 1.
See also
aminThe minimum value of a dataset along a given dimension, propagating NaNs.
minimumElement-wise minimum of two datasets, propagating any NaNs.
maximumElement-wise maximum of two datasets, propagating any NaNs.
fmaxElement-wise maximum of two datasets, ignoring any NaNs.
fminElement-wise minimum of two datasets, ignoring any NaNs.
argmaxReturn the indices or coordinates of the maximum values.
argminReturn the indices or coordinates of the minimum values.
Notes
For dataset with complex or hypercomplex type type, the default is the value with the maximum real part.
- mean(dataset, dim=None, dtype=None, keepdims=False)[source]
Compute the arithmetic mean along the specified axis.
Returns the average of the array elements. The average is taken over the flattened array by default, otherwise over the specified axis.
- Parameters:
dataset (array_like) – Array containing numbers whose mean is desired.
dim (None or int or dimension name, optional) – Dimension or dimensions along which to operate.
dtype (data-type, optional) – Type to use in computing the mean. For integer inputs, the default is
float64; for floating point inputs, it is the same as the input dtype.keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
mean – A new array containing the mean values.
See also
Notes
The arithmetic mean is the sum of the elements along the axis divided by the number of elements.
Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.mean(nd) <Quantity(1.25085858, 'absorbance')> >>> scp.mean(nd, keepdims=True) NDDataset: [float64] a.u. (shape: (y:1, x:1)) >>> m = scp.mean(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.x Coord: [float64] cm⁻¹ (size: 5549)
- min(dataset, dim=None, keepdims=False, **kwargs)[source]
Return the maximum of the dataset or maxima along given dimensions.
- Parameters:
dataset (array_like) – Input array or object that can be converted to an array.
dim (None or int or dimension name or tuple of int or dimensions, optional) – Dimension or dimensions along which to operate. By default, flattened input is used. If this is a tuple, the minimum is selected over multiple dimensions, instead of a single dimension or all the dimensions as before.
keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
amin – Minimum of the data. If
dimis None, the result is a scalar value. Ifdimis given, the result is an array of dimensionndim - 1.
See also
amaxThe maximum value of a dataset along a given dimension, propagating NaNs.
minimumElement-wise minimum of two datasets, propagating any NaNs.
maximumElement-wise maximum of two datasets, propagating any NaNs.
fmaxElement-wise maximum of two datasets, ignoring any NaNs.
fminElement-wise minimum of two datasets, ignoring any NaNs.
argmaxReturn the indices or coordinates of the maximum values.
argminReturn the indices or coordinates of the minimum values.
- ones(shape, dtype=None, **kwargs)[source]
Return a new
NDDatasetof given shape and type, filled with ones.- Parameters:
shape (int or sequence of ints) – Shape of the new array, e.g.,
(2, 3)or2.dtype (data-type, optional) – The desired data-type for the array, e.g.,
numpy.int8. Default is**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
ones – Array of
ones.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
zeros_likeReturn an array of zeros with shape and type of input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
full_likeFill an array with shape and type of input.
zerosReturn a new array setting values to zero.
emptyReturn a new uninitialized array.
fullFill a new array.
Examples
>>> nd = scp.ones(5, units='km') >>> nd NDDataset: [float64] km (size: 5) >>> nd.values <Quantity([ 1 1 1 1 1], 'kilometer')> >>> nd = scp.ones((5,), dtype=np.int, mask=[True, False, False, False, True]) >>> nd NDDataset: [int64] unitless (size: 5) >>> nd.values masked_array(data=[ --, 1, 1, 1, --], mask=[ True, False, False, False, True], fill_value=999999) >>> nd = scp.ones((5,), dtype=np.int, mask=[True, False, False, False, True], units='joule') >>> nd NDDataset: [int64] J (size: 5) >>> nd.values <Quantity([ -- 1 1 1 --], 'joule')> >>> scp.ones((2, 2)).values array([[ 1, 1], [ 1, 1]])
- ones_like(dataset, dtype=None, **kwargs)[source]
Return
NDDatasetof ones.The returned
NDDatasethave the same shape and type as a given array. Units, coordset, … can be added in kwargs.- Parameters:
dataset (
NDDatasetor array-like) – Object from which to copy the array structure.dtype (data-type, optional) – Overrides the data type of the result.
**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
oneslike – Array of
1with the same shape and type asdataset.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
full_likeReturn an array with a given fill value with shape and type of the
input.zeros_likeReturn an array of zeros with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
zerosReturn a new array setting values to zero.
onesReturn a new array setting values to one.
emptyReturn a new uninitialized array.
fullFill a new array.
Examples
>>> x = np.arange(6) >>> x = x.reshape((2, 3)) >>> x = scp.NDDataset(x, units='s') >>> x NDDataset: [float64] s (shape: (y:2, x:3)) >>> scp.ones_like(x, dtype=float, units='J') NDDataset: [float64] J (shape: (y:2, x:3))
- pipe(func, *args, **kwargs)[source]
Apply func(self, *args, **kwargs).
- Parameters:
func (function) – Function to apply to the
NDDataset. *args, and **kwargs are passed intofunc. Alternatively a(callable, data_keyword)tuple wheredata_keywordis a string indicating the keyword ofcallablethat expects the array object.*args – Positional arguments passed into
func.**kwargs – Keyword arguments passed into
func.
- Returns:
pipe – The return type of
func.
Notes
Use
pipewhen chaining together functions that expect aNDDataset.
- plot(method=None, **kwargs)[source]
Plot the dataset using the specified method.
- Parameters:
dataset (
NDDataset) – Source of data to plot.method (str, optional, default:
preference.method_1Dorpreference.method_2D) – Name of plotting method to use. If None, method is chosen based on data dimensionality.1D plotting methods:
pen: Solid line plotbar: Bar graphscatter: Scatter plotscatter+pen: Scatter plot with solid line
2D plotting methods:
**kwargs (keyword parameters, optional) – See Other Parameters.
- Other Parameters:
ax (Axe, optional) – Axe where to plot. If not specified, create a new one.
clear (bool, optional, default: True) – If false, hold the current figure and ax until a new plot is performed.
color or c (color, optional, default: auto) – color of the line.
colorbar (bool, optional, default: True) – Show colorbar (2D plots only).
commands (str,) – matplotlib commands to be executed.
data_only (bool, optional, default: False) – Only the plot is done. No addition of axes or label specifications.
dpi (int, optional) – the number of pixel per inches.
figsize (tuple, optional, default is (3.4, 1.7)) – figure size.
fontsize (int, optional) – The font size in pixels, default is 10 (or read from preferences).
imag (bool, optional, default: False) – Show imaginary component for complex data. By default the real component is displayed.
linestyle or ls (str, optional, default: auto) – line style definition.
linewidth or lw (float, optional, default: auto) – line width.
marker, m (str, optional, default: auto) – marker type for scatter plot. If marker != “” then the scatter type of plot is chosen automatically.
markeredgecolor or mec (color, optional)
markeredgewidth or mew (float, optional)
markerfacecolor or mfc (color, optional)
markersize or ms (float, optional)
markevery (None or int)
modellinestyle or modls (str) – line style of the model.
offset (float) – offset of the model individual lines.
output (str,) – name of the file to save the figure.
plot_model (Bool,) – plot model data if available.
plottitle (bool, optional, default: False) – Use the name of the dataset as title. Works only if title is not defined
projections (bool, optional, default: False) – Show projections on the axes (2D plots only).
reverse (bool or None [optional, default=None/False) – In principle, coordinates run from left to right, except for wavenumbers (e.g., FTIR spectra) or ppm (e.g., NMR), that spectrochempy will try to guess. But if reverse is set, then this is the setting which will be taken into account.
show_complex (bool, optional, default: False) – Show both real and imaginary component for complex data. By default only the real component is displayed.
show_mask (bool, optional) – Should we display the mask using colored area.
show_z (bool, optional, default: True) – should we show the vertical axis.
show_zero (bool, optional) – show the zero basis.
style (str, optional, default:
scp.preferences.style(scpy)) – Matplotlib stylesheet (useavailable_styleto get a list of available styles for plotting.title (str) – Title of the plot (or subplot) axe.
transposed (bool, optional, default: False) – Transpose the data before plotting (2D plots only).
twinx (
Axesinstance, optional, default: None) – If this is not None, then a twin axes will be created with a common x dimension.uselabel_x (bool, optional) – use x coordinate label as x tick labels
vshift (float, optional) – vertically shift the line from its baseline.
xlim (tuple, optional) – limit on the horizontal axis.
xlabel (str, optional) – label on the horizontal axis.
x_reverse (bool, optional, default: False) – reverse the x axis. Equivalent to
reverse.ylabel or zlabel (str, optional) – label on the vertical axis.
ylim or zlim (tuple, optional) – limit on the vertical axis.
y_reverse (bool, optional, default: False) – reverse the y axis (2D plot only).
- Returns:
Matplolib Axes or None – The matplotlib axes containing the plot if successful, None otherwise.
- ptp(dataset, dim=None, keepdims=False)[source]
Range of values (maximum - minimum) along a dimension.
The name of the function comes from the acronym for ‘peak to peak’ .
- Parameters:
dim (None or int or dimension name, optional) – Dimension along which to find the peaks. If None, the operation is made on the first dimension.
keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input dataset.
- Returns:
ptp – A new dataset holding the result.
- random(size=None, dtype=None, **kwargs)[source]
Return random floats in the half-open interval [0.0, 1.0).
Results are from the “continuous uniform” distribution over the stated interval.
Note
To sample \(\\mathrm{Uniform}[a, b)\) with \(b > a\), multiply the output of random by (b-a) and add a, i.e.: \((b - a) * \mathrm{random}() + a\).
- Parameters:
size (int or tuple of ints, optional) – Output shape. If the given shape is, e.g., (m, n, k), then m * n * k samples are drawn. Default is None, in which case a single value is returned.
dtype (dtype, optional) – Desired dtype of the result, only float64 and float32 are supported. The default value is np.float64.
**kwargs – Keywords argument used when creating the returned object, such as units, name, title, etc…
- Returns:
random – Array of random floats of shape size (unless size=None, in which case a single float is returned).
- round(dataset, decimals=0)[source]
Evenly round to the given number of decimals.
- Parameters:
dataset (
NDDataset) – Input dataset.decimals (int, optional) – Number of decimal places to round to (default: 0). If decimals is negative, it specifies the number of positions to the left of the decimal point.
- Returns:
rounded_array – NDDataset containing the rounded values. The real and imaginary parts of complex numbers are rounded separately. The result of rounding a float is a float. If the dataset contains masked data, the mask remain unchanged.
See also
numpy.round,around,spectrochempy.round,spectrochempy.around,methods.,ceil,fix,floor,rint,trunc
- round_(dataset, decimals=0)[source]
Evenly round to the given number of decimals.
- Parameters:
dataset (
NDDataset) – Input dataset.decimals (int, optional) – Number of decimal places to round to (default: 0). If decimals is negative, it specifies the number of positions to the left of the decimal point.
- Returns:
rounded_array – NDDataset containing the rounded values. The real and imaginary parts of complex numbers are rounded separately. The result of rounding a float is a float. If the dataset contains masked data, the mask remain unchanged.
See also
numpy.round,around,spectrochempy.round,spectrochempy.around,methods.,ceil,fix,floor,rint,trunc
- save(**kwargs: Any)[source]
Save dataset in native .scp format.
- Parameters:
**kwargs (Any) – Optional arguments passed to save_as()
- Returns:
Optional[pathlib.Path] – Path to saved file if successful, None if save failed
- save_as(filename: str = "", **kwargs: Any) -> pathlib.Path | None: """ Save the current NDDataset in SpectroChemPy format (.scp). Parameters ---------- filename : str The filename of the file where to save the current dataset. **kwargs Optional keyword parameters (see Other Parameters). Other Parameters ---------------- directory : str, optional If specified, the given `directory` and the `filename` will be appended. See Also -------- save : Save current dataset. write : Export current dataset to different format. Notes ----- Adapted from :class:`numpy.savez` . Examples -------- Read some data from an OMNIC file >>> nd = scp.read_omnic('wodger.spg') >>> assert nd.name == 'wodger' Write it in SpectroChemPy format (.scp) (return a `pathlib` object) >>> filename = nd.save_as('new_wodger') Check the existence of the scp file >>> assert filename.is_file() >>> assert filename.name == 'new_wodger.scp' Remove this file >>> filename.unlink() """ if filename: # we have a filename # by default it use the saved directory filename = pathclean(filename) if self.directory and self.directory != filename.parent: filename = self.directory / filename else: filename = self.directory # suffix must be specified which correspond to the type of the # object to save default_suffix = SCPY_SUFFIX[self._implements()] if filename is not None and not filename.is_dir()[source]
Save the current NDDataset in SpectroChemPy format (.scp).
- Parameters:
filename (str) – The filename of the file where to save the current dataset.
**kwargs – Optional keyword parameters (see Other Parameters).
- Other Parameters:
directory (str, optional) – If specified, the given
directoryand thefilenamewill be appended.
Notes
Adapted from
numpy.savez.Examples
Read some data from an OMNIC file
>>> nd = scp.read_omnic('wodger.spg') >>> assert nd.name == 'wodger'
Write it in SpectroChemPy format (.scp) (return a
pathlibobject)>>> filename = nd.save_as('new_wodger')
Check the existence of the scp file
>>> assert filename.is_file() >>> assert filename.name == 'new_wodger.scp'
Remove this file
>>> filename.unlink()
- set_complex(inplace=False)[source]
Set the object data as complex.
When nD-dimensional array are set to complex, we assume that it is along the first dimension. Two succesives rows are merged to form a complex rows. This means that the number of row must be even If the complexity is to be applied in other dimension, either transpose/swapdims your data before applying this function in order that the complex dimension is the first in the array.
- Parameters:
inplace (bool, optional, default=False) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDComplexArray – Same object or a copy depending on the
inplaceflag.
See also
- set_coordset(*args, **kwargs)[source]
Set one or more coordinates at once.
- Parameters:
Warning
This method replace all existing coordinates.
See also
add_coordsetAdd one or a set of coordinates from a dataset.
set_coordtitlesSet titles of the one or more coordinates.
set_coordunitsSet units of the one or more coordinates.
- sort(**kwargs)[source]
Return the dataset sorted along a given dimension.
By default, it is the last dimension [axis=-1]) using the numeric or label values.
- Parameters:
dim (str or int, optional, default=-1) – Dimension index or name along which to sort.
pos (int , optional) – If labels are multidimensional - allow to sort on a define row of labels : labels[pos]. Experimental : Not yet checked.
by (str among [‘value’, ‘label’], optional, default=`value`) – Indicate if the sorting is following the order of labels or numeric coord values.
descend (
bool, optional, default=`False`) – If true the dataset is sorted in a descending direction. Default is False except if coordinates are reversed.inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDDataset– Sorted dataset.
- squeeze(*dims, inplace=False)[source]
Remove single-dimensional entries from the shape of a NDDataset.
- Parameters:
*dims (None or int or tuple of ints, optional) – Selects a subset of the single-dimensional entries in the shape. If a dimension (dim) is selected with shape entry greater than one, an error is raised.
inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDDataset– The input array, but with all or a subset of the dimensions of length 1 removed.- Raises:
ValueError – If
dimis notNone, and the dimension being squeezed is not of length 1.
- std(dataset, dim=None, dtype=None, ddof=0, keepdims=False)[source]
Compute the standard deviation along the specified axis.
Returns the standard deviation, a measure of the spread of a distribution, of the array elements. The standard deviation is computed for the flattened array by default, otherwise over the specified axis.
- Parameters:
dataset (array_like) – Calculate the standard deviation of these values.
dim (None or int or dimension name , optional) – Dimension or dimensions along which to operate. By default, flattened input is used.
dtype (dtype, optional) – Type to use in computing the standard deviation. For arrays of integer type the default is float64, for arrays of float types it is the same as the array type.
ddof (int, optional) – Means Delta Degrees of Freedom. The divisor used in calculations is
N - ddof, whereNrepresents the number of elements. By defaultddofis zero.keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
std – A new array containing the standard deviation.
Notes
The standard deviation is the square root of the average of the squared deviations from the mean, i.e.,
std = sqrt(mean(abs(x - x.mean())**2)).The average squared deviation is normally calculated as
x.sum() / N, whereN = len(x). If, however,ddofis specified, the divisorN - ddofis used instead. In standard statistical practice,ddof=1provides an unbiased estimator of the variance of the infinite population.ddof=0provides a maximum likelihood estimate of the variance for normally distributed variables. The standard deviation computed in this function is the square root of the estimated variance, so even withddof=1, it will not be an unbiased estimate of the standard deviation per se.Note that, for complex numbers,
stdtakes the absolute value before squaring, so that the result is always real and nonnegative. For floating-point input, the std is computed using the same precision the input has. Depending on the input data, this can cause the results to be inaccurate, especially for float32 (see example below). Specifying a higher-accuracy accumulator using thedtypekeyword can alleviate this issue.Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.std(nd) <Quantity(0.807972021, 'absorbance')> >>> scp.std(nd, keepdims=True) NDDataset: [float64] a.u. (shape: (y:1, x:1)) >>> m = scp.std(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.data array([ 0.08521, 0.08543, ..., 0.251, 0.2537])
- sum(dataset, dim=None, dtype=None, keepdims=False)[source]
Sum of array elements over a given axis.
- Parameters:
dataset (array_like) – Calculate the sum of these values.
dim (None or int or dimension name , optional) –
- Dimension or dimensions along which to operate. By default, flattened input
is used.
dtype (dtype, optional) – Type to use in computing the standard deviation. For arrays of integer type the default is float64, for arrays of float types it is the same as the array type.
keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
sum – A new array containing the sum.
See also
meanCompute the arithmetic mean along the specified axis.
trapzIntegration of array values using the composite trapezoidal rule.
Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.sum(nd) <Quantity(381755.783, 'absorbance')> >>> scp.sum(nd, keepdims=True) NDDataset: [float64] a.u. (shape: (y:1, x:1)) >>> m = scp.sum(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.data array([ 100.7, 100.7, ..., 74, 73.98])
- swapdims(dim1, dim2, inplace=False)[source]
Interchange two dimensions of a NDDataset.
- Parameters:
dim1 (int) – First axis.
dim2 (int) – Second axis.
inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDDataset– Swaped dataset.
See also
transposeTranspose a dataset.
- take(indices, **kwargs)[source]
Take elements from an array.
- Returns:
NDDataset– A sub dataset defined by the input indices.
- to(other, inplace=False, force=False)[source]
Return the object with data rescaled to different units.
- Parameters:
other (
Quantityor str) – Destination units.inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
force (bool, optional, default=False) – If True the change of units is forced, even for incompatible units.
- Returns:
rescaled
See also
itoInplace rescaling of the current object data to different units.
to_base_unitsRescaling of the current object data to different units.
ito_base_unitsInplace rescaling of the current object data to different units.
to_reduced_unitsRescaling to reduced_units.
ito_reduced_unitsInplace rescaling to reduced units.
Examples
>>> np.random.seed(12345) >>> ndd = scp.NDArray(data=np.random.random((3, 3)), ... mask=[[True, False, False], ... [False, True, False], ... [False, False, True]], ... units='meters') >>> print(ndd) NDArray: [float64] m (shape: (y:3, x:3))
We want to change the units to seconds for instance but there is no relation with meters, so an error is generated during the change
>>> ndd.to('second') Traceback (most recent call last): ... pint.errors.DimensionalityError: Cannot convert from 'meter' ([length]) to 'second' ([time])
However, we can force the change
>>> ndd.to('second', force=True) NDArray: [float64] s (shape: (y:3, x:3))
By default the conversion is not done inplace, so the original is not modified :
>>> print(ndd) NDArray: [float64] m (shape: (y:3, x:3))
- to_array()[source]
Return a numpy masked array.
Other NDDataset attributes are lost.
- Returns:
ndarray– The numpy masked array from the NDDataset data.
Examples
>>> dataset = scp.read('wodger.spg') >>> a = scp.to_array(dataset)
equivalent to:
>>> a = np.ma.array(dataset)
or
>>> a = dataset.masked_data
- to_base_units(inplace=False)[source]
Return an array rescaled to base units.
- Parameters:
inplace (bool) – If True the rescaling is done in place.
- Returns:
rescaled – A rescaled array.
- to_reduced_units(inplace=False)[source]
Return an array scaled in place to reduced units.
Reduced units means one unit per dimension. This will not reduce compound units (e.g., ‘J/kg’ will not be reduced to m**2/s**2).
- Parameters:
inplace (bool) – If True the rescaling is done in place.
- Returns:
rescaled – A rescaled array.
- to_xarray()[source]
Convert a NDDataset instance to an
DataArrayobject.Warning: the xarray library must be available.
- Returns:
object – A axrray.DataArray object.
- transpose(*dims, inplace=False)[source]
Permute the dimensions of a NDDataset.
- Parameters:
*dims (sequence of dimension indexes or names, optional) – By default, reverse the dimensions, otherwise permute the dimensions according to the values given.
inplace (bool, optional, default=`False`) – Flag to say that the method return a new object (default) or not (inplace=True).
- Returns:
NDDataset – Transposed NDDataset.
See also
swapdimsInterchange two dimensions of a NDDataset.
- var(dataset, dim=None, dtype=None, ddof=0, keepdims=False)[source]
Compute the variance along the specified axis.
Returns the variance of the array elements, a measure of the spread of a distribution. The variance is computed for the flattened array by default, otherwise over the specified axis.
- Parameters:
dataset (array_like) – Array containing numbers whose variance is desired.
dim (None or int or dimension name , optional) – Dimension or dimensions along which to operate. By default, flattened input is used.
dtype (dtype, optional) – Type to use in computing the standard deviation. For arrays of integer type the default is float64, for arrays of float types it is the same as the array type.
ddof (int, optional) – Means Delta Degrees of Freedom. The divisor used in calculations is
N - ddof, whereNrepresents the number of elements. By defaultddofis zero.keepdims (bool, optional) – If this is set to True, the dimensions which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
var – A new array containing the standard deviation.
See also
Notes
The variance is the average of the squared deviations from the mean, i.e.,
var = mean(abs(x - x.mean())**2).The mean is normally calculated as
x.sum() / N, whereN = len(x). If, however,ddofis specified, the divisorN - ddofis used instead. In standard statistical practice,ddof=1provides an unbiased estimator of the variance of a hypothetical infinite population.ddof=0provides a maximum likelihood estimate of the variance for normally distributed variables.Note that for complex numbers, the absolute value is taken before squaring, so that the result is always real and nonnegative.
For floating-point input, the variance is computed using the same precision the input has. Depending on the input data, this can cause the results to be inaccurate, especially for
float32(see example below). Specifying a higher-accuracy accumulator using thedtypekeyword can alleviate this issue.Examples
>>> nd = scp.read('irdata/nh4y-activation.spg') >>> nd NDDataset: [float64] a.u. (shape: (y:55, x:5549)) >>> scp.var(nd) <Quantity(0.652818786, 'absorbance')> >>> scp.var(nd, keepdims=True) NDDataset: [float64] a.u. (shape: (y:1, x:1)) >>> m = scp.var(nd, dim='y') >>> m NDDataset: [float64] a.u. (size: 5549) >>> m.data array([0.007262, 0.007299, ..., 0.06298, 0.06438])
- zeros(shape, dtype=None, **kwargs)[source]
Return a new
NDDatasetof given shape and type, filled with zeros.- Parameters:
shape (int or sequence of ints) – Shape of the new array, e.g.,
(2, 3)or2.dtype (data-type, optional) – The desired data-type for the array, e.g.,
numpy.int8. Default isnumpy.float64.**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
zeros – Array of zeros.
- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
zeros_likeReturn an array of zeros with shape and type of input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
full_likeFill an array with shape and type of input.
onesReturn a new array setting values to 1.
emptyReturn a new uninitialized array.
fullFill a new array.
Examples
>>> nd = scp.NDDataset.zeros(6) >>> nd NDDataset: [float64] unitless (size: 6) >>> nd = scp.zeros((5, )) >>> nd NDDataset: [float64] unitless (size: 5) >>> nd.values array([ 0, 0, 0, 0, 0]) >>> nd = scp.zeros((5, 10), dtype=np.int, units='absorbance') >>> nd NDDataset: [int64] a.u. (shape: (y:5, x:10))
- zeros_like(dataset, dtype=None, **kwargs)[source]
Return a
NDDatasetof zeros.The returned
NDDatasethave the same shape and type as a given array. Units, coordset, … can be added in kwargs.- Parameters:
dataset (
NDDatasetor array-like) – Object from which to copy the array structure.dtype (data-type, optional) – Overrides the data type of the result.
**kwargs – Optional keyword parameters (see Other Parameters).
- Returns:
zeorslike – Array of
fill_valuewith the same shape and type asdataset.- Other Parameters:
units (str or ur instance) – Units of the returned object. If not provided, try to copy from the input object.
coordset (list or Coordset object) – Coordinates for the returned object. If not provided, try to copy from the input object.
See also
full_likeReturn an array with a fill value with shape and type of the input.
ones_likeReturn an array of ones with shape and type of input.
empty_likeReturn an empty array with shape and type of input.
zerosReturn a new array setting values to zero.
onesReturn a new array setting values to one.
emptyReturn a new uninitialized array.
fullFill a new array.
Examples
>>> x = np.arange(6) >>> x = x.reshape((2, 3)) >>> nd = scp.NDDataset(x, units='s') >>> nd NDDataset: [float64] s (shape: (y:2, x:3)) >>> nd.values <Quantity([[ 0 1 2] [ 3 4 5]], 'second')> >>> nd = scp.zeros_like(nd) >>> nd NDDataset: [float64] s (shape: (y:2, x:3)) >>> nd.values <Quantity([[ 0 0 0] [ 0 0 0]], 'second')>
Examples using spectrochempy.NDDataset
Using plot_multiple to plot several datasets on the same figure
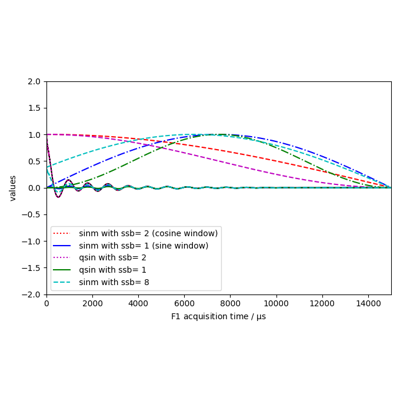
Sine bell and squared Sine bell window multiplication
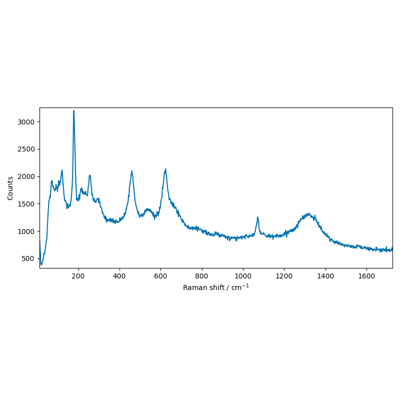
Savitky-Golay and Whittaker-Eilers smoothing of a Raman spectrum
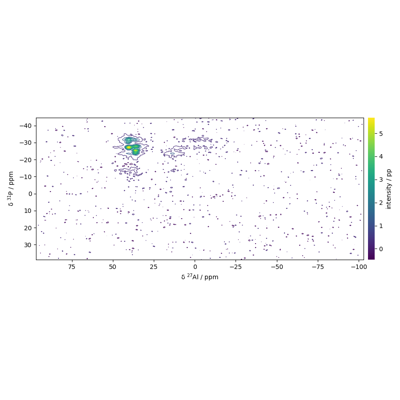
Processing NMR spectra (slicing, baseline correction, peak picking, peak fitting)