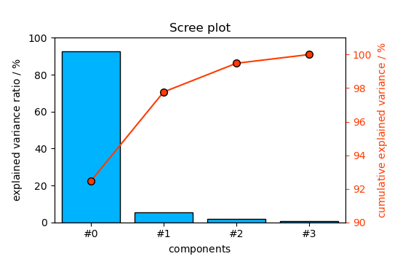
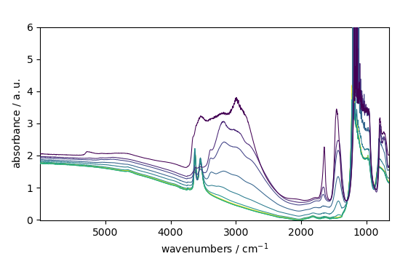
spectrochempy.PCA.scoreplot¶
- PCA.scoreplot(self, *pcs, colormap='viridis', color_mapping='index', show_labels=False, labels_column=0, labels_every=1, **kwargs)[source]¶
2D or 3D scoreplot of samples.
- Parameters
*pcs (a series of int argument or a list/tuple) – Must contain 2 or 3 elements.
colormap (str) – A matplotlib colormap.
color_mapping (‘index’ or ‘labels’) – If ‘index’, then the colors of each n_scores is mapped sequentially on the colormap. If labels, the labels of the n_observations are used for color mapping.
show_labels (bool, optional, default: False) – If True each observation will be annotated with its label.
labels_column (int, optional, default:0) – If several columns of labels are present indicates which column has to be used to show labels.
labels_every (int, optional, default: 1)
` Do not label all points, but only every value indicated by this parameter.