Note
Go to the end to download the full example code
NDDataset PCA analysis example¶
In this example, we perform the PCA dimensionality reduction of a spectra dataset
Import the spectrochempy API package
import spectrochempy as scp
Load a dataset
dataset = scp.read_omnic("irdata/nh4y-activation.spg")
dataset = dataset[:, 2000.0:4000.0] # remember float number to slice from coordinates
print(dataset)
dataset.plot_stack()
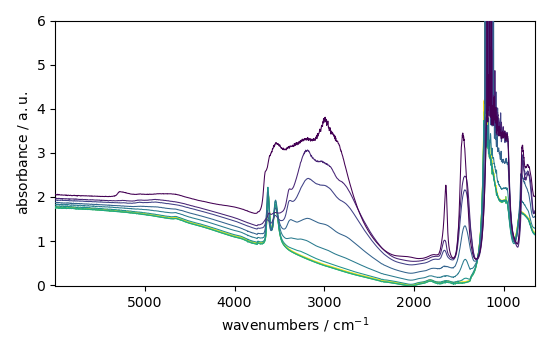
NDDataset: [float64] a.u. (shape: (y:55, x:2075))
<_Axes: xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
Create a PCA object
Reduce the dataset to a lower dimensionality (number of components is automatically determined)
S, LT = pca.reduce(n_pc=0.99)
print(LT)
NDDataset: [float64] a.u. (shape: (y:2, x:2075))
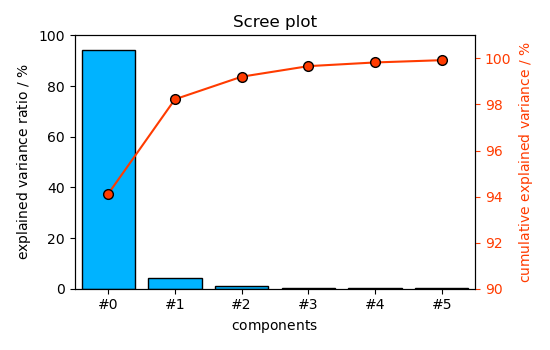
Finally, display the results graphically ScreePlot
_ = pca.screeplot()
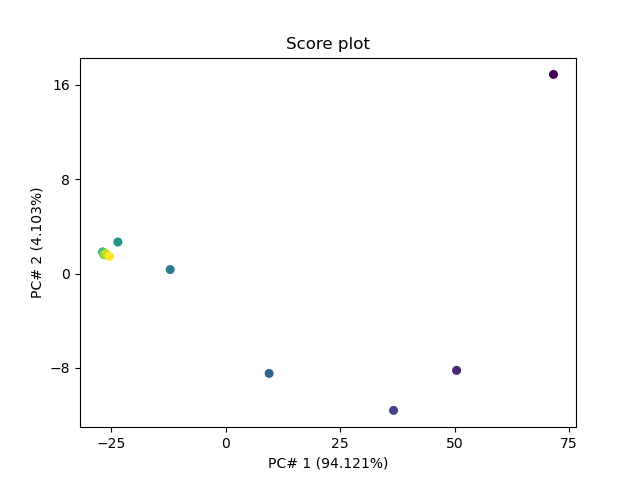
Score Plot
_ = pca.scoreplot(1, 2)
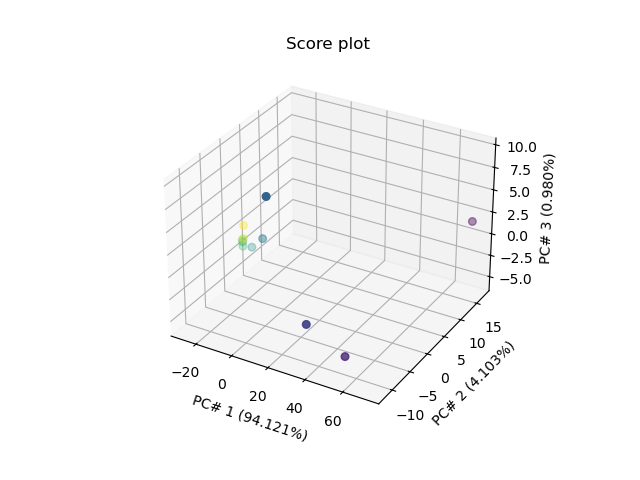
Score Plot for 3 PC’s in 3D
_ = pca.scoreplot(1, 2, 3)
Our dataset has already two columns of labels for the spectra but there are little too long for display on plots.
array([[ 2016-07-06 19:03:14+00:00, vz0466.spa, Wed Jul 06 21:00:38 2016 (GMT+02:00)],
[ 2016-07-06 19:13:14+00:00, vz0467.spa, Wed Jul 06 21:10:38 2016 (GMT+02:00)],
...,
[ 2016-07-07 04:03:17+00:00, vz0520.spa, Thu Jul 07 06:00:41 2016 (GMT+02:00)],
[ 2016-07-07 04:13:17+00:00, vz0521.spa, Thu Jul 07 06:10:41 2016 (GMT+02:00)]], dtype=object)
So we define some short labels for each component, and add them as a third column:
labels = [lab[:6] for lab in dataset.y.labels[:, 1]]
# we cannot change directly the label as S is read-only, but use the method `labels`
pca.labels(labels) # Note this does not replace previous labels, but adds a column.
now display thse
_ = pca.scoreplot(1, 2, show_labels=True, labels_column=2, labels_every=5)
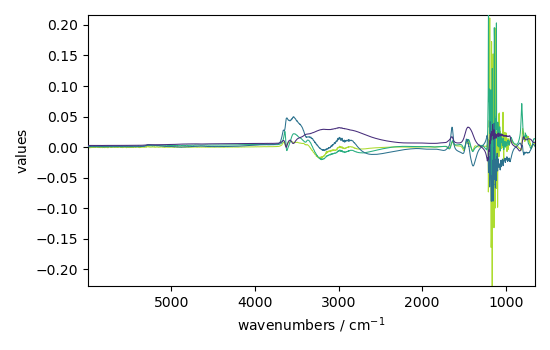
Displays the 4-first loadings
Total running time of the script: ( 0 minutes 1.896 seconds)