Note
Go to the end to download the full example code.
Fitting 1D dataset
In this example, we find the least square solution of a simple linear equation.
import os
import spectrochempy as scp
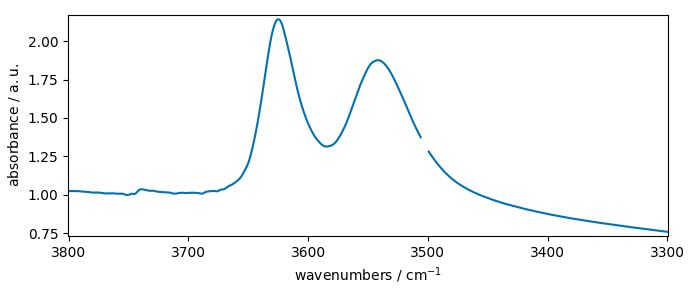
Let’s take an IR spectrum
nd = scp.read_omnic(os.path.join("irdata", "nh4y-activation.spg"))
and select only the OH region:
Perform a Fit Fit parameters are defined in a script (a single text as below)
script = """
#-----------------------------------------------------------
# syntax for parameters definition:
# name: value, low_bound, high_bound
# available prefix:
# # for comments
# * for fixed parameters
# $ for variable parameters
# > for reference to a parameter in the COMMON block
# (> is forbidden in the COMMON block)
# common block parameters should not have a _ in their names
#-----------------------------------------------------------
#
COMMON:
# common parameters ex.
# $ gwidth: 1.0, 0.0, none
$ gratio: 0.1, 0.0, 1.0
MODEL: LINE_1
shape: asymmetricvoigtmodel
* ampl: 1.1, 0.0, none
$ pos: 3620, 3400.0, 3700.0
$ ratio: 0.0147, 0.0, 1.0
$ asym: 0.1, 0, 1
$ width: 50, 0, 1000
MODEL: LINE_2
shape: asymmetricvoigtmodel
$ ampl: 0.8, 0.0, none
$ pos: 3540, 3400.0, 3700.0
> ratio: gratio
$ asym: 0.1, 0, 1
$ width: 50, 0, 1000
"""
create an Optimize object
f1 = scp.Optimize(log_level="INFO")
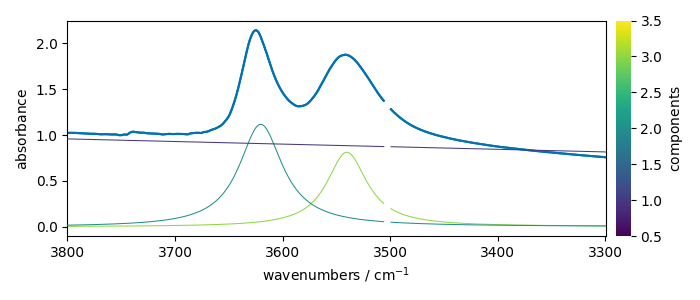
Show plot and the starting model using the dry parameters (of course it is advisable to be as close as possible of a good expectation
f1.script = script
# set dry and continue to show starting model
# reset dry and continue to show starting model
f1.dry = True
f1.autobase = True
f1.fit(ndOH)
# get some information
scp.info_(f"numbers of components: {f1.n_components}")
ndOH.plot()
ax = (f1.components[:]).plot(clear=False)
ax.autoscale(enable=True, axis="y")
**************************************************
Starting parameters:
**************************************************
COMMON:
$ gratio: 0.1000, 0.0, 1.0
MODEL: line_1
shape: asymmetricvoigtmodel
* ampl: 1.1000, 0.0, none
$ asym: 0.1000, 0, 1
$ pos: 3620.0000, 3400.0, 3700.0
$ ratio: 0.0147, 0.0, 1.0
$ width: 50.0000, 0, 1000
MODEL: line_2
shape: asymmetricvoigtmodel
$ ampl: 0.8000, 0.0, none
$ asym: 0.1000, 0, 1
$ pos: 3540.0000, 3400.0, 3700.0
> ratio:gratio
$ width: 50.0000, 0, 1000
numbers of components: 2
Now perform a fit with maximum 1000 iterations
f1.max_iter = 1000
f1.fit(ndOH)
**************************************************
Result:
**************************************************
COMMON:
$ gratio: 0.3458, 0.0, 1.0
MODEL: line_1
shape: asymmetricvoigtmodel
* ampl: 1.1000, 0.0, none
$ asym: 0.7716, 0, 1
$ pos: 3623.4044, 3400.0, 3700.0
$ ratio: 0.4394, 0.0, 1.0
$ width: 43.5995, 0, 1000
MODEL: line_2
shape: asymmetricvoigtmodel
$ ampl: 0.9001, 0.0, none
$ asym: 1.0000, 0, 1
$ pos: 3536.9977, 3400.0, 3700.0
> ratio:gratio
$ width: 79.4888, 0, 1000
<spectrochempy.analysis.curvefitting.optimize.Optimize object at 0x7f3ab19d7950>
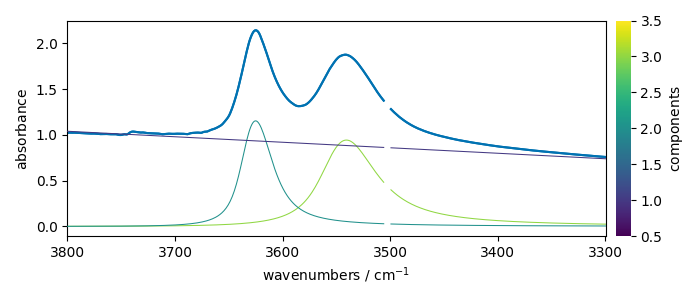
Show the result
ndOH.plot()
ax = (f1.components[:]).plot(clear=False)
ax.autoscale(enable=True, axis="y")
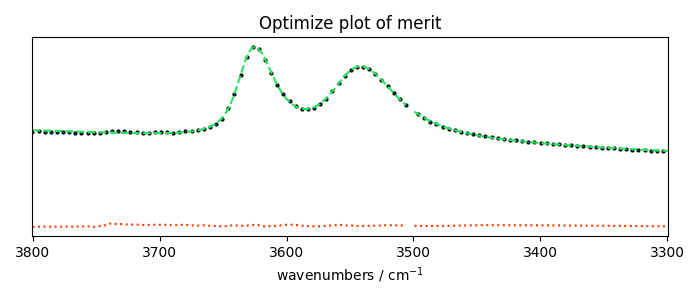
plotmerit
som = f1.inverse_transform()
f1.plotmerit(ndOH, som, method="scatter", markevery=5, markersize=2, lw=2)
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: (0 minutes 0.855 seconds)