spectrochempy.em
- em(lb="Hz", shifted="us")def em(dataset, lb=1, shifted=0, **kwargs)[source]
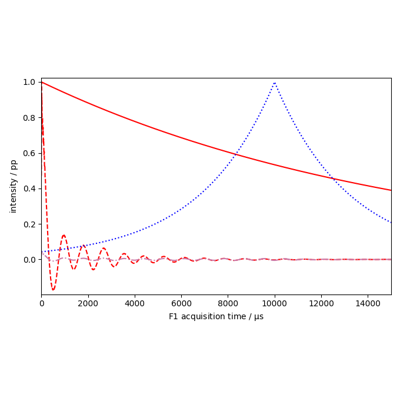
Calculate exponential apodization.
For multidimensional NDDataset, the apodization is by default performed on the last dimension.
The data in the last dimension MUST be time-domain, or an error is raised.
Functional form of apodization window :
\[em(t) = \exp(- e (t-t_0) )\]where
\[e = \pi * lb\]- Parameters:
dataset (Dataset) – Input dataset.
lb (float or
Quantity, optional, default=1 Hz) – Exponential line broadening, If it is not a quantity with units, it is assumed to be a broadening expressed in Hz.shifted (float or
quantity, optional, default=0 us) – Shift the data time origin by this amount. If it is not a quantity it is assumed to be expressed in the data units of the last dimension.
- Returns:
apodized – Dataset.
apod_arr – The apodization array only if ‘retapod’ is True.
- Other Parameters:
dim (str or int, keyword parameter, optional, default=’x’) – Specify on which dimension to apply the apodization method. If
dimis specified as an integer it is equivalent to the usualaxisnumpy parameter.inv (bool, keyword parameter, optional, default=False) – True for inverse apodization.
rev (bool, keyword parameter, optional, default=False) – True to reverse the apodization before applying it to the data.
inplace (bool, keyword parameter, optional, default=False) – True if we make the transform inplace. If False, the function return a new dataset.
retapod (bool, keyword parameter, optional, default=False) – True to return the apodization array along with the apodized object.