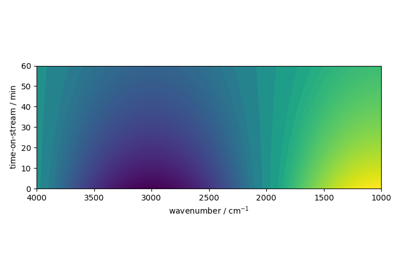
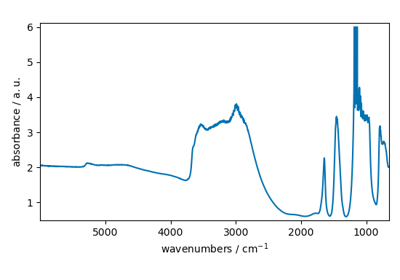
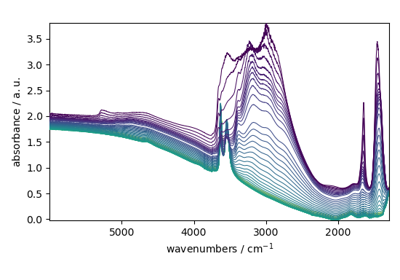
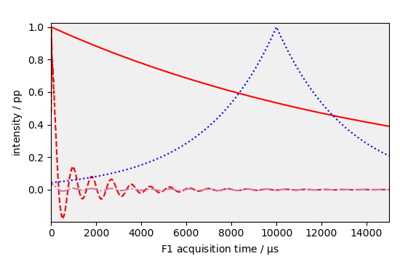
spectrochempy.NDDataset¶
- class NDDataset(**kwargs)[source]¶
The main N-dimensional dataset class used by SpectroChemPy .
The NDDataset is the main object use by SpectroChemPy. Like numpy ndarrays, NDDataset have the capability to be sliced, sorted and subject to mathematical operations. But, in addition, NDDataset may have units, can be masked and each dimensions can have coordinates also with units. This make NDDataset aware of unit compatibility, e.g., for binary operation such as additions or subtraction or during the application of mathematical operations. In addition or in replacement of numerical data for coordinates, NDDataset can also have labeled coordinates where labels can be different kind of objects (strings, datetime, numpy nd.ndarray or other NDDatasets, etc…).
- Parameters
data (array of floats) – Data array contained in the object. The data can be a list, a tuple, a
ndarray, a ndarray-like, aNDArrayor any subclass ofNDArray. Any size or shape of data is accepted. If not given, an emptyNDArraywill be inited. At the initialisation the provided data will be eventually casted to a numpy-ndarray. If a subclass ofNDArrayis passed which already contains some mask, labels, or units, these elements will be used to accordingly set those of the created object. If possible, the provided data will not be copied fordatainput, but will be passed by reference, so you should make a copy of thedatabefore passing them if that’s the desired behavior or set thecopyargument to True.coordset (An instance of |CoordSet| , optional) –
coordscontains the coordinates for the different dimensions of thedata. ifcoordsis provided, it must specified thecoordandlabelsfor all dimensions of thedata. Multiplecoord’s can be specified in anCoordSetinstance for each dimension.coordunits (list, optional) – A list of units corresponding to the dimensions in the order of the coordset.
coordtitles (list, optional) – A list of titles corresponding of the dimensions in the order of the coordset.
**kwargs – Optional keyword parameters (see Other Parameters).
- Other Parameters
dtype (str or dtype, optional, default=np.float64) – If specified, the data will be cast to this dtype, else the data will be cast to float64 or complex128.
dims (list of chars, optional) – If specified the list must have a length equal to the number od data dimensions (ndim) and the chars must be taken among x,y,z,u,v,w or t. If not specified, the dimension names are automatically attributed in this order.
name (str, optional) – A user-friendly name for this object. If not given, the automatic
idgiven at the object creation will be used as a name.labels (array of objects, optional) – Labels for the
data. labels can be used only for 1D-datasets. The labels array may have an additional dimension, meaning several series of labels for the same data. The given array can be a list, a tuple, andarray, a ndarray-like, aNDArrayor any subclass ofNDArray.mask (array of bool or
NOMASK, optional) – Mask for the data. The mask array must have the same shape as the data. The given array can be a list, a tuple, or andarray. Each values in the array must beFalsewhere the data are valid and True when they are not (like in numpy masked arrays). Ifdatais already aMaskedArray, or any array object (such as aNDArrayor subclass of it), providing amaskhere, will causes the mask from the masked array to be ignored.units (|Unit| instance or str, optional) – Units of the data. If data is a
Quantitythenunitsis set to the unit of thedata; if a unit is also explicitly provided an error is raised. Handling of units use the pint package.timezone (datetime.tzinfo, optional) – The timezone where the data were created. If not specified, the local timezone is assumed.
title (str, optional) – The title of the data dimension. The
titleattribute should not be confused with thename. Thetitleattribute is used for instance for labelling plots of the data. It is optional but recommended to give a title to each ndarray data.dlabel (str, optional) – Alias of
title.meta (dict-like object, optional) – Additional metadata for this object. Must be dict-like but no further restriction is placed on meta.
author (str, optional) – Name(s) of the author(s) of this dataset. BNy default, name of the computer note where this dataset is created.
description (str, optional) – An optional description of the nd-dataset. A shorter alias is
desc.origin (str, optional) – Origin of the data: Name of organization, address, telephone number, name of individual contributor, etc., as appropriate.
roi (list) – Region of interest (ROI) limits.
history (str, optional) – A string to add to the object history.
copy (bool, optional) – Perform a copy of the passed object. Default is False.
See also
CoordExplicit coordinates object.
LinearCoordImplicit coordinates object.
CoordSetSet of coordinates.
Notes
The underlying array in a
NDDatasetobject can be accessed through thedataattribute, which will return a conventionalndarray.Methods
NDDataset.ab(dataset[, dim])Alias of
abc.NDDataset.abc(dataset[, dim])Automatic baseline correction.
NDDataset.abs(dataset[, dtype])Calculate the absolute value of the given NDDataset element-wise.
NDDataset.absolute(dataset[, dtype])Calculate the absolute value of the given NDDataset element-wise.
NDDataset.add_coordset(*coords[, dims])Add one or a set of coordinates from a dataset.
NDDataset.align(dataset, *others, **kwargs)Align individual
NDDatasetalong given dimensions using various methods.NDDataset.all(dataset[, dim, keepdims])Test whether all array elements along a given axis evaluate to True.
NDDataset.amax(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
NDDataset.amin(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
NDDataset.any(dataset[, dim, keepdims])Test whether any array element along a given axis evaluates to True.
NDDataset.arange([start, stop, step, dtype])Return evenly spaced values within a given interval.
NDDataset.argmax(dataset[, dim])Indexes of maximum of data along axis.
NDDataset.argmin(dataset[, dim])Indexes of minimum of data along axis.
NDDataset.around(dataset[, decimals])Evenly round to the given number of decimals.
Make data and mask (ndim >= 1) laid out in Fortran order in memory.
NDDataset.astype([dtype])Cast the data to a specified type.
NDDataset.autosub(dataset, ref, *ranges[, ...])Automatic subtraction of a reference to the dataset.
NDDataset.average(dataset[, dim, weights, ...])Compute the weighted average along the specified axis.
NDDataset.bartlett()def bartlett(dataset, ...)Calculate Bartlett apodization (triangular window with end points at zero).
NDDataset.basc(dataset, *ranges, **kwargs)Compute a baseline correction using the BaselineCorrection processor.
Calculate a minimum 4-term Blackman-Harris apodization.
NDDataset.clip(dataset[, a_min, a_max])Clip (limit) the values in a dataset.
Close a Matplotlib figure associated to this dataset.
NDDataset.component([select])Take selected components of an hypercomplex array (RRR, RIR, ...).
NDDataset.concatenate(*datasets, **kwargs)Concatenation of
NDDatasetobjects along a given axis.NDDataset.conj(dataset[, dim])Conjugate of the NDDataset in the specified dimension.
NDDataset.conjugate(dataset[, dim])Conjugate of the NDDataset in the specified dimension.
NDDataset.coord([dim])Return the coordinates along the given dimension.
NDDataset.coordmax(dataset[, dim])Find coordinates of the maximum of data along axis.
NDDataset.coordmin(dataset[, dim])Find oordinates of the mainimum of data along axis.
NDDataset.copy([deep, keepname])Make a disconnected copy of the current object.
NDDataset.cs(dataset[, pts, neg])Circular shift.
NDDataset.cumsum(dataset[, dim, dtype])Return the cumulative sum of the elements along a given axis.
NDDataset.dc(dataset, **kwargs)Time domain baseline correction.
Delete all coordinate settings.
NDDataset.detrend(dataset[, type, bp])Remove linear trend along dim from dataset.
NDDataset.diag(dataset[, offset])Extract a diagonal or construct a diagonal array.
NDDataset.diagonal(dataset[, offset, dim, dtype])Return the diagonal of a 2D array.
NDDataset.dot(a, b[, strict, out])Return the dot product of two NDDatasets.
Upload the classical "iris" dataset.
NDDataset.download_nist_ir(CAS[, index])Upload IR spectra from NIST webbook
NDDataset.dump(filename, **kwargs)Save the current object into compressed native spectrochempy format.
NDDataset.em([lb, shifted, lb, shifted])Calculate exponential apodization.
NDDataset.empty(shape[, dtype])Return a new
NDDatasetof given shape and type, without initializing entries.NDDataset.empty_like(dataset[, dtype])Return a new uninitialized
NDDataset.NDDataset.expand_dims([dim])Expand the shape of an array.
NDDataset.eye(N[, M, k, dtype])Return a 2-D array with ones on the diagonal and zeros elsewhere.
NDDataset.fft(dataset[, size, sizeff, inv, ppm])Apply a complex fast fourier transform.
NDDataset.find_peaks(dataset[, height, ...])Wrapper and extension of scpy.signal.find_peaks().
NDDataset.fromfunction(cls, function[, ...])Construct a nddataset by executing a function over each coordinate.
NDDataset.fromiter(iterable[, dtype, count])Create a new 1-dimensional array from an iterable object.
NDDataset.fsh(dataset, pts, **kwargs)Frequency shift by Fourier transform.
NDDataset.fsh2(dataset, pts, **kwargs)Frequency Shift by Fourier transform.
NDDataset.full(shape[, fill_value, dtype])Return a new
NDDatasetof given shape and type, filled withfill_value.NDDataset.full_like(dataset[, fill_value, dtype])Return a
NDDatasetof fill_value.NDDataset.general_hamming([alpha])Calculate generalized Hamming apodization.
NDDataset.geomspace(start, stop[, num, ...])Return numbers spaced evenly on a log scale (a geometric progression).
NDDataset.get_axis(*args, **kwargs)Helper function to determine an axis index whatever the syntax used (axis index or dimension names).
NDDataset.get_labels([level])Get the labels at a given level.
NDDataset.gm([gb, lb, shifted, gb, lb, shifted])Calculate lorentz-to-gauss apodization.
NDDataset.hamming(dataset, **kwargs)Calculate generalized Hamming (== Happ-Genzel) apodization.
NDDataset.hann(dataset, **kwargs)Return a Hann window.
NDDataset.ht(dataset[, N])Hilbert transform.
NDDataset.identity(n[, dtype])Return the identity
NDDatasetof a given shape.NDDataset.ifft(dataset[, size])Apply a inverse fast fourier transform.
Check the compatibility of units with another object.
NDDataset.ito(other[, force])Inplace scaling to different units.
Inplace rescaling to base units.
Quantity scaled in place to reduced units, inplace.
NDDataset.linspace(cls, start, stop[, num, ...])Return evenly spaced numbers over a specified interval.
NDDataset.load(filename, **kwargs)Open data from a '*.scp' (NDDataset) or '.pscp' (Project) file.
NDDataset.logspace(cls, start, stop[, num, ...])Return numbers spaced evenly on a log scale.
NDDataset.ls(dataset[, pts])Left shift and zero fill.
NDDataset.max(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
NDDataset.mc(dataset)Modulus calculation.
NDDataset.mean(dataset[, dim, dtype, keepdims])Compute the arithmetic mean along the specified axis.
NDDataset.min(dataset[, dim, keepdims])Return the maximum of the dataset or maxima along given dimensions.
NDDataset.ones(shape[, dtype])Return a new
NDDatasetof given shape and type, filled with ones.NDDataset.ones_like(dataset[, dtype])Return
NDDatasetof ones.NDDataset.pipe(func, *args, **kwargs)NDDataset.pk(dataset[, phc0, phc1, exptc, pivot])Linear phase correction.
NDDataset.pk_exp(dataset[, phc0, pivot, exptc])Exponential Phase Correction.
NDDataset.plot([method])Generic plot function.
NDDataset.plot_1D(...[, method])Plot of one-dimensional data.
NDDataset.plot_2D(...[, method])Plot of 2D array.
NDDataset.plot_3D(...[, method])Plot of 2D array as 3D plot
NDDataset.plot_bar(dataset, *args, **kwargs)Plot a 1D dataset with bars.
NDDataset.plot_image(dataset, *args, **kwargs)Plot a 2D dataset as an image plot.
NDDataset.plot_map(dataset, *args, **kwargs)Plot a 2D dataset as a contoured map.
NDDataset.plot_multiple(datasets[, method, ...])Plot a series of 1D datasets as a scatter plot with optional lines between markers.
NDDataset.plot_pen(dataset, *args, **kwargs)Plot a 1D dataset with solid pen by default.
NDDataset.plot_scatter(dataset, *args, **kwargs)Plot a 1D dataset as a scatter plot (points can be added on lines).
NDDataset.plot_scatter_pen(dataset, *args, ...)Plot a 1D dataset with solid pen by default.
NDDataset.plot_stack(dataset, *args, **kwargs)Plot a 2D dataset as a stack plot.
NDDataset.plot_surface(dataset, *args, **kwargs)Plot a 2D dataset as a a 3D-surface.
NDDataset.plot_waterfall(dataset, *args, ...)Plot a 2D dataset as a a 3D-waterfall plot.
NDDataset.ps(dataset)Power spectrum.
NDDataset.ptp(dataset[, dim, keepdims])Range of values (maximum - minimum) along a dimension.
NDDataset.qsin(dataset[, ssb])Equivalent to
sp, with pow = 2 (squared sine bell apodization window).NDDataset.random([size, dtype])Return random floats in the half-open interval [0.0, 1.0).
NDDataset.read(*paths, **kwargs)Generic read method.
NDDataset.read_carroucell([directory])Open .spa files in a directory after a carroucell experiment.
NDDataset.read_csv(*paths, **kwargs)NDDataset.read_ddr(*paths, **kwargs)Open a Surface Optics Corps.
NDDataset.read_dir([directory])Read an entire directory.
NDDataset.read_hdr(*paths, **kwargs)Open a Surface Optics Corps.
NDDataset.read_jcamp(*paths, **kwargs)Open Infrared JCAMP-DX files with extension
.jdxor.dx.NDDataset.read_labspec(*paths, **kwargs)Read a single Raman spectrum or a series of Raman spectra.
NDDataset.read_mat(*paths, **kwargs)Read a matlab file with extension
.matand return its content as a list.NDDataset.read_matlab(*paths, **kwargs)Read a matlab file with extension
.matand return its content as a list.NDDataset.read_omnic(*paths, **kwargs)Open a Thermo Nicolet OMNIC file.
NDDataset.read_opus(*paths, **kwargs)Open Bruker OPUS file(s).
NDDataset.read_quadera(*paths, **kwargs)Read a Pfeiffer Vacuum's QUADERA® mass spectrometer software file with extension
.asc.NDDataset.read_remote(file_or_dir, **kwargs)Download and read files or an entire directory on github spectrochempy_data repository.
NDDataset.read_sdr(*paths, **kwargs)Open a Surface Optics Corps.
NDDataset.read_soc(*paths, **kwargs)Open a Surface Optics Corps.
NDDataset.read_spa(*paths, **kwargs)Open a Thermo Nicolet file or a list of files with extension
.spa.NDDataset.read_spc(*paths, **kwargs)Open a Thermo Nicolet file or a list of files with extension
.spg.NDDataset.read_spg(*paths, **kwargs)Open a Thermo Nicolet file or a list of files with extension
.spg.NDDataset.read_srs(*paths, **kwargs)Open a Thermo Nicolet file or a list of files with extension
.srs.NDDataset.read_topspin(*paths, **kwargs)Open Bruker TOPSPIN (NMR) dataset.
NDDataset.read_zip(*paths, **kwargs)Open a zipped list of data files.
Remove all masks previously set on this array.
NDDataset.roll(dataset[, pts, neg])Roll dimensions.
NDDataset.round(dataset[, decimals])Evenly round to the given number of decimals.
NDDataset.round_(dataset[, decimals])Evenly round to the given number of decimals.
NDDataset.rs(dataset[, pts])Right shift and zero fill.
NDDataset.save(**kwargs)Save the current object in SpectroChemPy format.
NDDataset.save_as([filename])NDDataset.savgol_filter(dataset[, ...])Apply a Savitzky-Golay filter to a NDDataset.
NDDataset.set_complex([inplace])Set the object data as complex.
NDDataset.set_coordset(*args, **kwargs)Set one or more coordinates at once.
NDDataset.set_coordtitles(*args, **kwargs)Set titles of the one or more coordinates.
NDDataset.set_coordunits(*args, **kwargs)Set units of the one or more coordinates.
NDDataset.set_hypercomplex([inplace])Alias of set_quaternion.
NDDataset.set_quaternion([inplace])Alias of set_quaternion.
NDDataset.simpson(dataset, *args, **kwargs)Integrate using the composite Simpson's rule.
NDDataset.sine(dataset, *args, **kwargs)Strictly equivalent to
sp.NDDataset.sinm(dataset[, ssb])Equivalent to
sp, with pow = 1 (sine bell apodization window).NDDataset.smooth(dataset[, window_length, ...])Smooth the data using a window with requested size.
NDDataset.sort(**kwargs)Return the dataset sorted along a given dimension.
NDDataset.sp([ssb, pow, ssb, pow])Calculate apodization with a Sine window multiplication.
NDDataset.squeeze(*dims[, inplace])Remove single-dimensional entries from the shape of a NDDataset.
NDDataset.stack(*datasets)Stack of
NDDatasetobjects along a new dimension.NDDataset.std(dataset[, dim, dtype, ddof, ...])Compute the standard deviation along the specified axis.
NDDataset.sum(dataset[, dim, dtype, keepdims])Sum of array elements over a given axis.
NDDataset.swapaxes(dim1, dim2[, inplace])Alias of
swapdims.NDDataset.swapdims(dim1, dim2[, inplace])Interchange two dimensions of a NDDataset.
NDDataset.take(indices, **kwargs)Take elements from an array.
NDDataset.to(other[, inplace, force])Return the object with data rescaled to different units.
Return a numpy masked array.
NDDataset.to_base_units([inplace])Return an array rescaled to base units.
NDDataset.to_reduced_units([inplace])Return an array scaled in place to reduced units.
Convert a NDDataset instance to an
DataArrayobject.NDDataset.transpose(*dims[, inplace])Permute the dimensions of a NDDataset.
NDDataset.trapezoid(dataset, **kwargs)Integrate using the composite trapezoidal rule.
NDDataset.trapz(*args, **kwargs)An alias of
trapezoidkept for backwards compatibility.NDDataset.triang()def triang(dataset, **kwargs)Calculate triangular apodization with non-null extremities and maximum value normalized to 1.
NDDataset.var(dataset[, dim, dtype, ddof, ...])Compute the variance along the specified axis.
NDDataset.write(dataset[, filename])Write the current dataset.
NDDataset.write_csv(*args, **kwargs)Write a dataset in CSV format.
NDDataset.write_excel(*args, **kwargs)This method is an alias of
write_excel.NDDataset.write_jcamp(*args, **kwargs)This method is an alias of
write_jcamp.NDDataset.write_jdx(*args, **kwargs)This method is an alias of
write_jcamp.NDDataset.write_mat(*args, **kwargs)This method is an alias of
write_matlab.NDDataset.write_matlab(*args, **kwargs)This method is an alias of
write_matlab.NDDataset.write_xls(*args, **kwargs)This method is an alias of
write_excel.NDDataset.zeros(shape[, dtype])Return a new
NDDatasetof given shape and type, filled with zeros.NDDataset.zeros_like(dataset[, dtype])Return a
NDDatasetof zeros.NDDataset.zf(dataset[, size, mid])Zero fill to given size.
NDDataset.zf_auto(dataset[, mid])Zero fill to next largest power of two.
NDDataset.zf_double(dataset, n[, mid])Zero fill by doubling original data size once or multiple times.
NDDataset.zf_size(dataset[, size, mid])Zero fill to given size.
Attributes
The array with imaginary-imaginary component of hypercomplex 2D data.
The array with imaginary-real component of hypercomplex 2D
data.The array with real-imaginary component of hypercomplex 2D
data.The array with real component in both dimension of hypercomplex 2D
data.Transposed
NDDataset.Creator of the dataset (str).
The main matplotlib axe associated to this dataset.
The matplotlib axe associated to the transposed dataset.
Matplotlib colorbar axe associated to this dataset.
Matplotlib colorbar axe associated to the transposed dataset.
Matplotlib projection x axe associated to this dataset.
Matplotlib projection y axe associated to this dataset.
Provides a comment (Alias to the description attribute).
List of the
Coordnames.CoordSetinstance.List of the
Coordtitles.List of the
Coordunits.Creation date object (Datetime).
The
dataarray.Provides a description of the underlying data (str).
True if the
dataarray is dimensionless - Readonly property (bool).Names of the dimensions (list).
Current directory for this dataset.
Matplotlib plot divider.
Data type (np.dtype).
Matplotlib figure associated to this dataset.
Matplotlib figure associated to this dataset.
Current filename for this dataset.
Type of current file.
True if at least one of the
dataarray dimension is complex.True if the
dataarray is not empty and size > 0.True is the name has been defined (bool).
True if the
dataarray have units - Readonly property (bool).Describes the history of actions made on this array (tr.List of strings).
Object identifier - Readonly property (str).
The array with imaginary component of the
data.True if the
dataarray has only one dimension (bool).True if the 'data' are complex (Readonly property).
True if the
dataarray is empty or size=0, and if no label are present.True if the
dataare real values - Readonly property (bool).True if the
dataare integer values - Readonly property (bool).True if the
dataarray is hypercomplex with interleaved data.True if the
dataarray have labels - Readonly property (bool).True if the
dataarray has masked values.True if the
dataarray is hypercomplex .Range of the data.
Return the local timezone.
Mask for the data (
ndarrayof bool).The actual masked
dataarray - Readonly property (array).Additional metadata (
Meta).ndarray- models data.Date of modification (readonly property).
A user-friendly name (str).
A dictionary containing all the axes of the current figures.
The number of dimensions of the
dataarray (Readonly property).Origin of the data.
Projectinstance.Metainstance object - Additional metadata.The array with real component of the
data.Region of interest (ROI) limits (list).
A tuple with the size of each dimension - Readonly property.
Size of the underlying
dataarray - Readonly property (int).Filename suffix.
Return the timezone information.
An user friendly title (str).
The actual array with mask and unit (
Quantity).bool- True if thedatadoes not haveunits(Readonly property).Unit- The units of the data.Alias of
values.Quantity- The actual values (data, units) contained in this object (Readonly property).- II¶
The array with imaginary-imaginary component of hypercomplex 2D data.
(Readonly property).
- RR¶
The array with real component in both dimension of hypercomplex 2D
data.This readonly property is equivalent to the
realproperty.
- author¶
Creator of the dataset (str).
- ax¶
The main matplotlib axe associated to this dataset.
- axT¶
The matplotlib axe associated to the transposed dataset.
- axec¶
Matplotlib colorbar axe associated to this dataset.
- axecT¶
Matplotlib colorbar axe associated to the transposed dataset.
- axex¶
Matplotlib projection x axe associated to this dataset.
- axey¶
Matplotlib projection y axe associated to this dataset.
- comment¶
Provides a comment (Alias to the description attribute).
- coordnames¶
List of the
Coordnames.Read only property.
- coordset¶
CoordSetinstance.Contains the coordinates of the various dimensions of the dataset. It’s a readonly property. Use set_coords to change one or more coordinates at once.
- coordtitles¶
List of the
Coordtitles.Read only property. Use set_coordtitle to eventually set titles.
- coordunits¶
List of the
Coordunits.Read only property. Use set_coordunits to eventually set units.
- created¶
Creation date object (Datetime).
- data¶
The
dataarray.If there is no data but labels, then the labels are returned instead of data.
- description¶
Provides a description of the underlying data (str).
- dims¶
Names of the dimensions (list).
The name of the dimensions are ‘x’, ‘y’, ‘z’…. depending on the number of dimension.
- directory¶
Current directory for this dataset.
ReadOnly property - automatically set when the filename is updated if it contains a parent on its path.
- divider¶
Matplotlib plot divider.
- dtype¶
Data type (np.dtype).
- fig¶
Matplotlib figure associated to this dataset.
- fignum¶
Matplotlib figure associated to this dataset.
- filename¶
Current filename for this dataset.
- filetype¶
Type of current file.
- has_defined_name¶
True is the name has been defined (bool).
- has_units¶
True if the
dataarray have units - Readonly property (bool).See also
unitlessTrue if the data has no unit.
dimensionlessTrue if the data have dimensionless units.
- history¶
Describes the history of actions made on this array (tr.List of strings).
- id¶
Object identifier - Readonly property (str).
- is_complex¶
True if the ‘data’ are complex (Readonly property).
- is_empty¶
True if the
dataarray is empty or size=0, and if no label are present.Readonly property (bool).
- labels¶
An array of labels for
data(ndarrayof str).An array of objects of any type (but most generally string), with the last dimension size equal to that of the dimension of data. Note that’s labelling is possible only for 1D data. One classical application is the labelling of coordinates to display informative strings instead of numerical values.
- limits¶
Range of the data.
- local_timezone¶
Return the local timezone.
- m¶
-
If there is no data but labels, then the labels are returned instead of data.
- magnitude¶
-
If there is no data but labels, then the labels are returned instead of data.
- meta¶
Additional metadata (
Meta).
- modified¶
Date of modification (readonly property).
- name¶
A user-friendly name (str).
When no name is provided, the
idof the object is returned instead.
- ndaxes¶
A dictionary containing all the axes of the current figures.
- origin¶
Origin of the data.
e.g. spectrometer or software
- parent¶
Projectinstance.The parent project of the dataset.
- preferences¶
Metainstance object - Additional metadata.
- roi¶
Region of interest (ROI) limits (list).
- shape¶
A tuple with the size of each dimension - Readonly property.
The number of
dataelement on each dimension (possibly complex). For only labelled array, there is no data, so it is the 1D and the size is the size of the array of labels.
- size¶
Size of the underlying
dataarray - Readonly property (int).The total number of data elements (possibly complex or hypercomplex in the array).
- suffix¶
Filename suffix.
Read Only property - automatically set when the filename is updated if it has a suffix, else give the default suffix for the given type of object.
- timezone¶
Return the timezone information.
A timezone’s offset refers to how many hours the timezone is from Coordinated Universal Time (UTC).
A
naivedatetime object contains no timezone information. The easiest way to tell if a datetime object is naive is by checking tzinfo. will be set to None of the object is naive.A naive datetime object is limited in that it cannot locate itself in relation to offset-aware datetime objects.
In spectrochempy, all datetimes are stored in UTC, so that conversion must be done during the display of these datetimes using tzinfo.
- title¶
An user friendly title (str).
When the title is provided, it can be used for labeling the object, e.g., axe title in a matplotlib plot.
- umasked_data¶
The actual array with mask and unit (
Quantity).(Readonly property).
- units¶
Unit- The units of the data.
- values¶
Quantity- The actual values (data, units) contained in this object (Readonly property).
Examples using spectrochempy.NDDataset¶
MCR-ALS optimization example (original example from Jaumot)
EFA analysis (Keller and Massart original example)
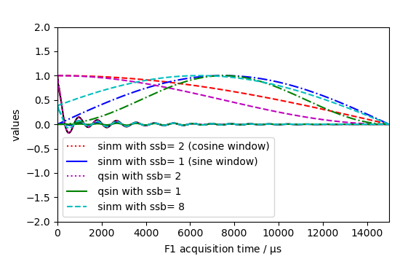
Sine bell and squared Sine bell window multiplication