Note
Go to the end to download the full example code.
EFA example
In this example, we perform the Evolving Factor Analysis
sphinx_gallery_thumbnail_number = 2
import os
import spectrochempy as scp
Upload and preprocess a dataset
datadir = scp.preferences.datadir
dataset = scp.read_omnic(os.path.join(datadir, "irdata", "nh4y-activation.spg"))
Change the time origin
columns masking
difference spectra dataset -= dataset[-1]
dataset.plot_stack(title="NH4_Y activation dataset")
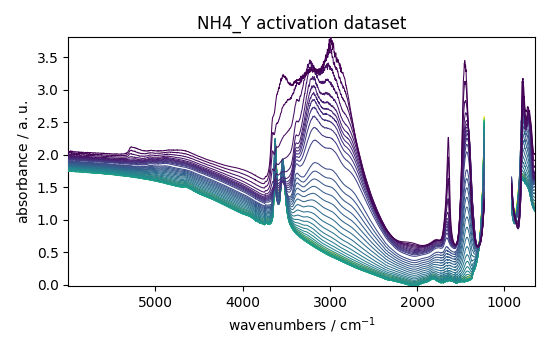
<_Axes: title={'center': 'NH4_Y activation dataset'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
Evolving Factor Analysis
Forward evolution of the 5 first components
f = efa1.f_ev[:, :5]
f.T.plot(yscale="log", legend=f.k.labels)
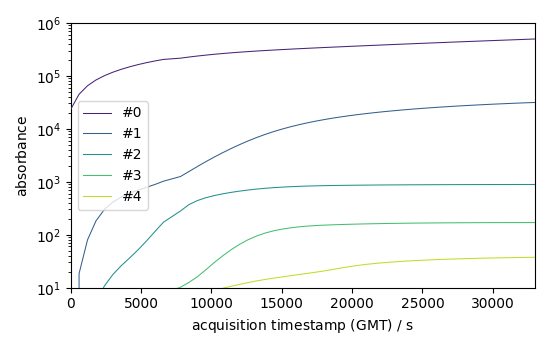
<_Axes: xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='absorbance $\\mathrm{}$'>
Note the use of coordinate ‘k’ (component axis) in the expression above.
Remember taht to find the actul names of the coordinates, the dims
attribute can be used as in the following:
f.dims
# Backward evolution
b = efa1.b_ev[:, :5]
b.T[:5].plot(yscale="log", legend=b.k.labels)
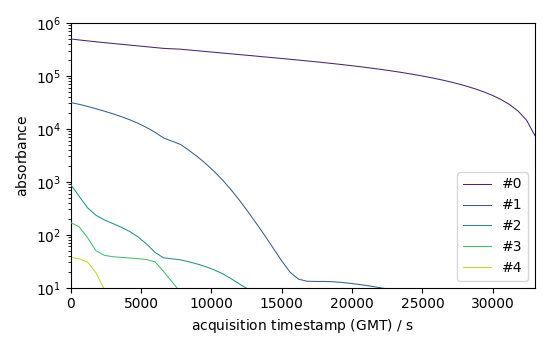
<_Axes: xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='absorbance $\\mathrm{}$'>
Show results with 3 components (which seems to already explain a large part of the dataset) we use the magnitude of the 4th component for the cut-off value (assuming it corresponds mostly to noise)
efa1.n_components = 3
efa1.cutoff = efa1.f_ev[:, 3].max()
# get concentration
C1 = efa1.transform()
C1.T.plot(title="EFA determined concentrations", legend=C1.k.labels)
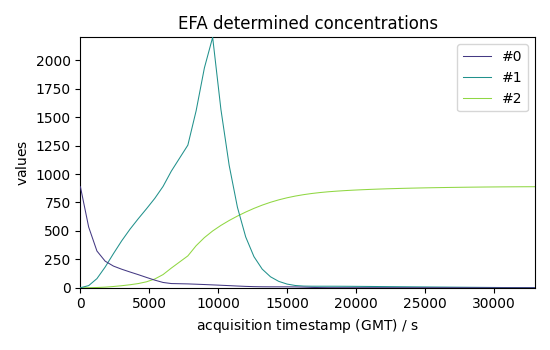
<_Axes: title={'center': 'EFA determined concentrations'}, xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='values $\\mathrm{}$'>
Fit transform : Get the concentration in too commands The number of desired components can be passed to the EFA model, followed by the fit_transform method:
Get components
St = efa2.get_components()
St.plot(title="components", legend=St.k.labels)
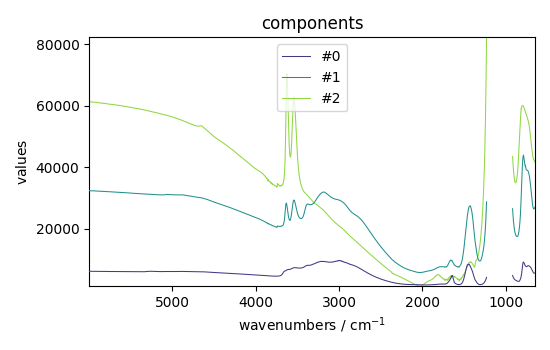
<_Axes: title={'center': 'components'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='values $\\mathrm{}$'>
Compare with PCA
pca = scp.PCA(n_components=3)
C3 = pca.fit_transform(dataset)
C3.T.plot(title="PCA scores")
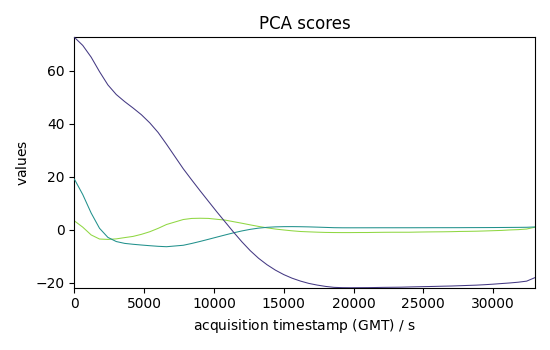
<_Axes: title={'center': 'PCA scores'}, xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='values $\\mathrm{}$'>
LT = pca.loadings
LT.plot(title="PCA components", legend=LT.k.labels)
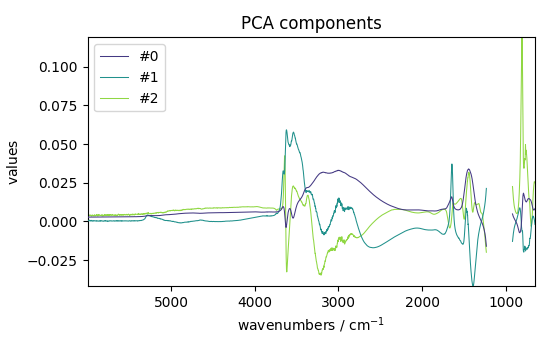
<_Axes: title={'center': 'PCA components'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='values $\\mathrm{}$'>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: (0 minutes 2.924 seconds)