Peak Integration
This tutorial shows how to find peak maxima and determine peak areas with SpectroChemPy. As a prerequisite, the user is expected to have read the Import, Import IR, Slicing, and Baseline Correction tutorials.
First, let’s import the SpectroChemPy API.
[1]:
import spectrochempy as scp
|
|
SpectroChemPy's API - v.0.7.1 © Copyright 2014-2025 - A.Travert & C.Fernandez @ LCS |
Now, import some 2D data into an NDDataset object.
[2]:
ds = scp.read_omnic("irdata/nh4y-activation.spg")
ds
[2]:
| name | nh4y-activation |
| author | runner@fv-az1774-299 |
| created | 2025-02-25 08:01:13+00:00 |
| description | Omnic title: NH4Y-activation.SPG Omnic filename: /home/runner/.spectrochempy/testdata/irdata/nh4y-activation.spg |
| history | 2025-02-25 08:01:13+00:00> Imported from spg file /home/runner/.spectrochempy/testdata/irdata/nh4y-activation.spg. 2025-02-25 08:01:13+00:00> Sorted by date |
| DATA | |
| title | absorbance |
| values | [[ 2.057 2.061 ... 2.013 2.012] [ 2.033 2.037 ... 1.913 1.911] ... [ 1.794 1.791 ... 1.198 1.198] [ 1.816 1.815 ... 1.24 1.238]] a.u. |
| shape | (y:55, x:5549) |
| DIMENSION `x` | |
| size | 5549 |
| title | wavenumbers |
| coordinates | [ 6000 5999 ... 650.9 649.9] cm⁻¹ |
| DIMENSION `y` | |
| size | 55 |
| title | acquisition timestamp (GMT) |
| coordinates | [1.468e+09 1.468e+09 ... 1.468e+09 1.468e+09] s |
| labels | [[ 2016-07-06 19:03:14+00:00 2016-07-06 19:13:14+00:00 ... 2016-07-07 04:03:17+00:00 2016-07-07 04:13:17+00:00] [ vz0466.spa, Wed Jul 06 21:00:38 2016 (GMT+02:00) vz0467.spa, Wed Jul 06 21:10:38 2016 (GMT+02:00) ... vz0520.spa, Thu Jul 07 06:00:41 2016 (GMT+02:00) vz0521.spa, Thu Jul 07 06:10:41 2016 (GMT+02:00)]] |
It’s a series of 55 spectra.
For the demonstration, select only the first 20 on a limited region from 1250 to 1800 cm\(^{-1}\) (Do not forget to use floating numbers for slicing).
[3]:
X = ds[:20, 1250.0:1800.0]
We can also eventually remove the offset on the acquisition time dimension (y).
[4]:
X.y -= X.y[0]
X.y.ito("min")
X.y.title = "acquisition time"
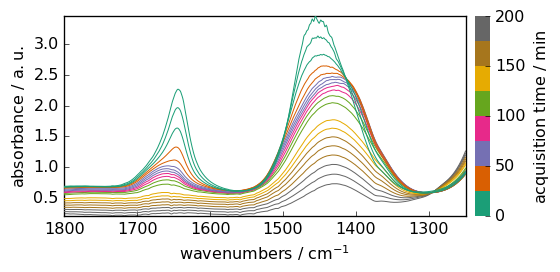
We set some plotting preferences and then plot the raw data.
[5]:
prefs = X.preferences
prefs.figure.figsize = (6, 3)
prefs.colormap = "Dark2"
prefs.colorbar = True
X.plot()
[5]:
<_Axes: xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
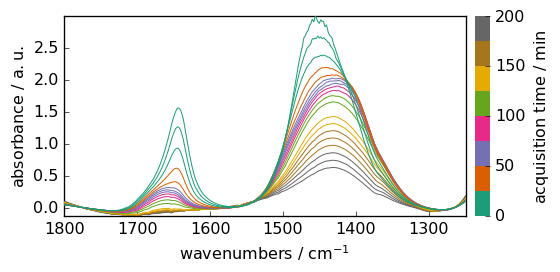
Now we can perform some baseline correction.
[6]:
blc = scp.Baseline()
blc.ranges = (
[1740.0, 1800.0],
[1550.0, 1570.0],
[1250.0, 1300.0],
) # define 3 regions where we want the baseline to reach zero.
blc.model = "polynomial"
blc.order = 3
blc.fit(X) # fit the baseline
Xcorr = blc.corrected # get the corrected dataset
Xcorr.plot()
[6]:
<_Axes: xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
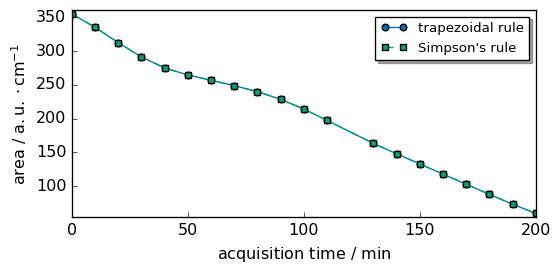
To integrate each row on the full range, we can use the sum or trapz method of an NDDataset.
[7]:
inttrapz = Xcorr.trapezoid(dim="x")
intsimps = Xcorr.simpson(dim="x")
As you can see, both methods give almost the same results in this case.
[8]:
scp.plot_multiple(
method="scatter",
ms=5,
datasets=[inttrapz, intsimps],
labels=["trapezoidal rule", "Simpson's rule"],
legend="best",
)
[8]:
<_Axes: xlabel='acquisition time $\\mathrm{/\\ \\mathrm{min}}$', ylabel='area $\\mathrm{/\\ \\mathrm{a.u.} \\cdot \\mathrm{cm}^{-1}}$'>
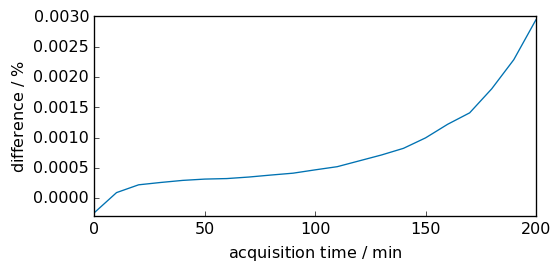
The difference between the trapezoidal and Simpson integration methods is visualized below. In this case, they are extremely close.
[9]:
diff = (inttrapz - intsimps) * 100.0 / intsimps
diff.title = "difference"
diff.units = "percent"
diff.plot(scatter=True, ms=5)
[9]:
<_Axes: xlabel='acquisition time $\\mathrm{/\\ \\mathrm{min}}$', ylabel='difference $\\mathrm{/\\ \\mathrm{\\%}}$'>