Project management
from pathlib import Path
[1]:
from spectrochempy import NDDataset
from spectrochempy import Project
from spectrochempy import pathclean
from spectrochempy import preferences as prefs
|
|
SpectroChemPy's API - v.0.7.1 © Copyright 2014-2025 - A.Travert & C.Fernandez @ LCS |
Project creation
We can easily create a new project to store various datasets
[2]:
proj = Project()
As we did not specify a name, a name has been attributed automatically:
[3]:
proj.name
[3]:
'Project-Project_c94b923e'
To get the signature of the object, one can use the usual ‘?’. Uncomment the following line to check
[4]:
# Project?
Let’s change this name
[5]:
proj.name = "myNMRdata"
proj
[5]:
(empty project)
Now we will add a dataset to the project.
First we read the dataset (here some NMR data) and we give it some name (e.g. ‘nmr n°1’)
[6]:
datadir = pathclean(prefs.datadir)
path = datadir / "nmrdata" / "bruker" / "tests" / "nmr"
nd1 = NDDataset.read_topspin(
path / "topspin_1d", expno=1, remove_digital_filter=True, name="NMR_1D"
)
nd2 = NDDataset.read_topspin(
path / "topspin_2d", expno=1, remove_digital_filter=True, name="NMR_2D"
)
WARNING | (UserWarning) (196608,)cannot be shaped into(147, 1024)
To add it to the project, we use the add_dataset function for a single dataset:
[7]:
proj.add_datasets(nd1)
or add_datasets for several datasets.
[8]:
proj.add_datasets(nd1, nd2)
Display its structure
[9]:
proj
[9]:
⤷ NMR_1D (dataset)
⤷ NMR_2D (dataset)
It is also possible to add other projects as sub-project (using the add_project )
Remove an element from a project
[10]:
proj.remove_dataset("NMR_1D")
proj
[10]:
⤷ NMR_2D (dataset)
Get project’s elements
[11]:
proj.add_datasets(nd1, nd2)
proj
[11]:
⤷ NMR_1D (dataset)
⤷ NMR_2D (dataset)
We can just use the name of the element as a project attribute.
[12]:
proj.NMR_1D
[12]:
| name | NMR_1D |
| author | runner@fv-az1774-299 |
| created | 2025-02-25 08:01:58+00:00 |
| DATA | |
| title | intensity |
| values | R[ 1078 2284 ... 0.2342 -0.1008] pp I[ -1037 -2200 ... 0.06203 -0.05273] pp |
| size | 12411 (complex) |
| DIMENSION `x` | |
| size | 12411 |
| title | F1 acquisition time |
| coordinates | [ 0 4 ... 4.964e+04 4.964e+04] µs |
[13]:
_ = proj.NMR_1D.plot()
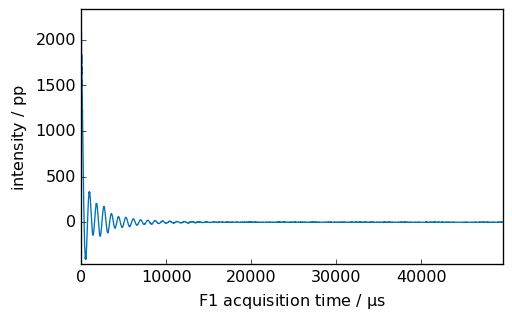
However, this work only if the name contains no space, dot, comma, colon, etc. The only special character allowed is the underscore _ . If the name is not respecting this, then it is possible to use the following syntax (as a project behave as a dictionary). For example:
[14]:
proj["NMR_1D"].data
[14]:
array([1.08e+03-1.04e+03j, 2.28e+03-2.2e+03j, ..., 0.234+0.062j, -0.101-0.0527j])
[15]:
proj.NMR_2D
[15]:
| name | NMR_2D |
| author | runner@fv-az1774-299 |
| created | 2025-02-25 08:01:58+00:00 |
| DATA | |
| title | intensity |
| values | RR[[ -0.2238 -0.1985 ... -0.1662 0.03262] [-0.006566 0.0282 ... -0.02949 -0.06717] ... [ 0 0 ... 0 0] [ 0 0 ... 0 0]] pp RI[[ 0.06219 0.1467 ... 0.04565 0.03068] [-0.05969 -0.08752 ... -0.05134 -0.05994] ... [ -0 -0 ... -0 -0] [ -0 -0 ... -0 -0]] pp IR[[ -0.1623 -0.0563 ... 0.02654 -0.01094] [ 0.1344 -0.006515 ... -0.08239 0.00516] ... [ 0 0 ... 0 0] [ 0 0 ... 0 0]] pp II[[-0.003312 -0.001535 ... 0.02067 -0.08058] [-0.05685 0.1174 ... 0.05831 -0.003414] ... [ -0 -0 ... -0 -0] [ -0 -0 ... -0 -0]] pp |
| shape | (y:96(complex), x:948(complex)) |
| DIMENSION `x` | |
| size | 948 |
| title | F2 acquisition time |
| coordinates | [ 0 48 ... 4.541e+04 4.546e+04] µs |
| DIMENSION `y` | |
| size | 96 |
| title | F1 acquisition time |
| coordinates | [ 0 74 ... 6956 7030] µs |
Saving and loading projects
[16]:
proj
[16]:
⤷ NMR_1D (dataset)
⤷ NMR_2D (dataset)
Saving
[17]:
proj.save_as("NMR")
[17]:
PosixPath('/home/runner/work/spectrochempy/spectrochempy/docs/sources/userguide/objects/project/NMR.pscp')
Loading
[18]:
proj2 = Project.load("NMR")
[19]:
proj2
[19]:
⤷ NMR_1D (dataset)
⤷ NMR_2D (dataset)
[20]:
_ = proj2.NMR_1D.plot()
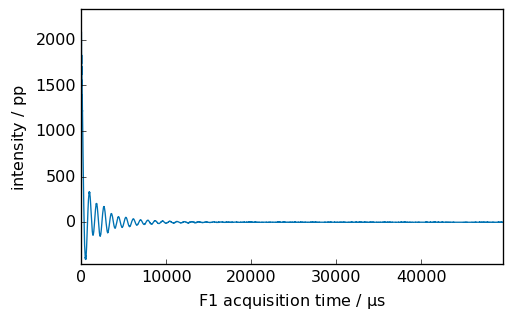
[21]:
proj2.NMR_2D
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File ~/work/spectrochempy/spectrochempy/.venv/lib/python3.10/site-packages/IPython/core/formatters.py:406, in BaseFormatter.__call__(self, obj)
404 method = get_real_method(obj, self.print_method)
405 if method is not None:
--> 406 return method()
407 return None
408 else:
File ~/work/spectrochempy/spectrochempy/src/spectrochempy/core/dataset/baseobjects/ndarray.py:857, in NDArray._repr_html_(self)
856 def _repr_html_(self):
--> 857 return convert_to_html(self)
File ~/work/spectrochempy/spectrochempy/src/spectrochempy/utils/print.py:112, in convert_to_html(obj)
102 tr = (
103 "<tr>"
104 "<td style='padding-right:5px; padding-bottom:0px; "
(...)
107 "padding-top:0px; {2} '>{1}</td><tr>\n"
108 )
110 obj._html_output = True
--> 112 out = obj._cstr()
114 regex = r"\0{3}[\w\W]*?\0{3}"
116 # noinspection PyPep8
File ~/work/spectrochempy/spectrochempy/src/spectrochempy/core/dataset/nddataset.py:592, in NDDataset._cstr(self)
588 out += f" author: {self.author}\n"
589 out += f" created: {self.created}\n"
590 out += (
591 f" modified: {self.modified}\n"
--> 592 if (self._modified - self._created).seconds > 30
593 else ""
594 )
596 wrapper1 = textwrap.TextWrapper(
597 initial_indent="",
598 subsequent_indent=" " * 15,
599 replace_whitespace=True,
600 width=self._text_width,
601 )
603 pars = self.description.strip().splitlines()
TypeError: can't subtract offset-naive and offset-aware datetimes
[21]:
NDDataset: [quaternion] pp (shape: (y:96, x:948))
[22]:
_ = proj.NMR_2D.plot()
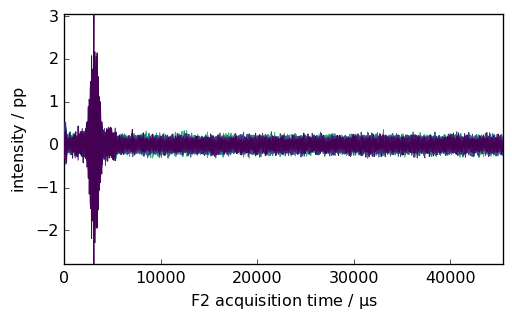
[ ]: