Note
Go to the end to download the full example code
EFA example¶
In this example, we perform the Evolving Factor Analysis
sphinx_gallery_thumbnail_number = 2
import os
import spectrochempy as scp
Upload and preprocess a dataset
datadir = scp.preferences.datadir
dataset = scp.read_omnic(os.path.join(datadir, "irdata", "nh4y-activation.spg"))
Change the time origin
columns masking
difference spectra dataset -= dataset[-1]
dataset.plot_stack(title="NH4_Y activation dataset")
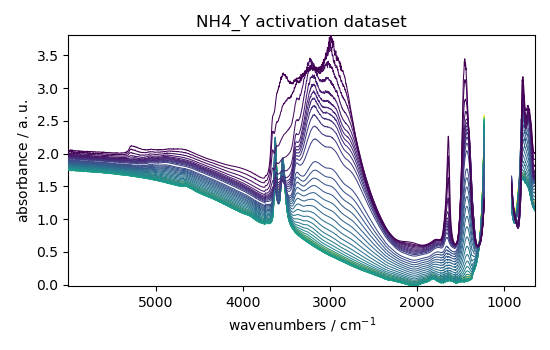
<_Axes: title={'center': 'NH4_Y activation dataset'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
Evolving Factor Analysis
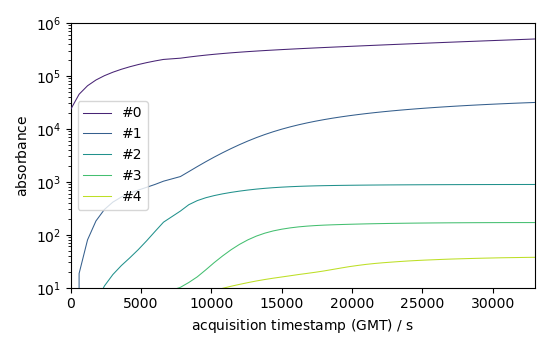
Forward evolution of the 5 first components
f = efa1.f_ev[:, :5]
f.T.plot(yscale="log", legend=f.x.labels)
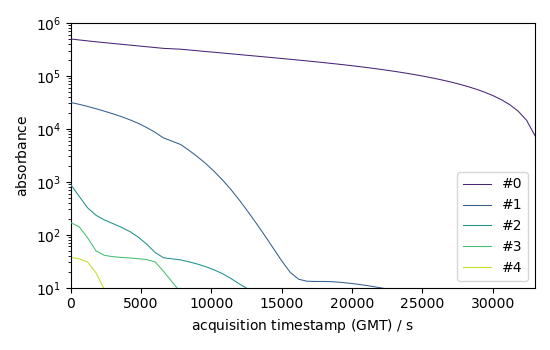
# Backward evolution
b = efa1.b_ev[:, :5]
b.T[:5].plot(yscale="log", legend=b.x.labels)
<_Axes: xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='absorbance $\\mathrm{}$'>
Show results with 3 components (which seems to already explain a large part of the dataset) we use the magnitude of the 4th component for the cut-off value (assuming it corresponds mostly to noise)
efa1.n_components = 3
efa1.cutoff = efa1.f_ev[:, 3].max()
# get concentration
C1 = efa1.transform()
C1.T.plot(title="EFA determined concentrations", legend=C1.x.labels)
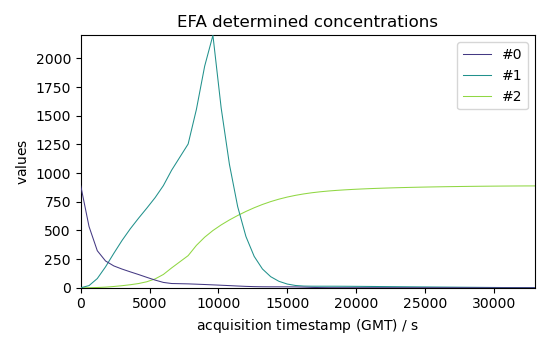
<_Axes: title={'center': 'EFA determined concentrations'}, xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='values $\\mathrm{}$'>
Fit transform : Get the concentration in too commands The number of desired components can be passed to the EFA model, followed by the fit_transform method:
Get components
St = efa2.get_components()
St.plot(title="components", legend=St.y.labels)
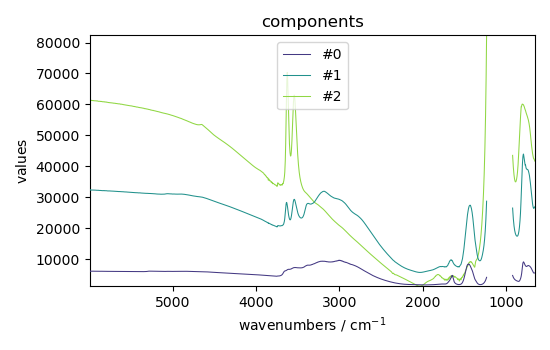
<_Axes: title={'center': 'components'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='values $\\mathrm{}$'>
Compare with PCA
pca = scp.PCA(n_components=3)
C3 = pca.fit_transform(dataset)
C3.T.plot(title="PCA scores")
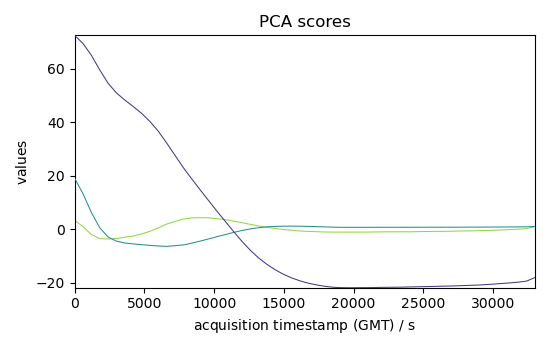
<_Axes: title={'center': 'PCA scores'}, xlabel='acquisition timestamp (GMT) $\\mathrm{/\\ \\mathrm{s}}$', ylabel='values $\\mathrm{}$'>
LT = pca.loadings
LT.plot(title="PCA components", legend=LT.y.labels)
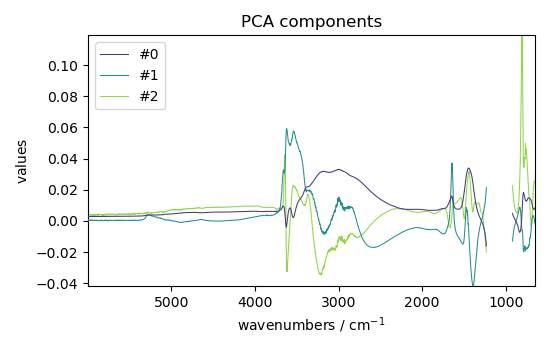
<_Axes: title={'center': 'PCA components'}, xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='values $\\mathrm{}$'>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script
# scp.show()
Total running time of the script: ( 0 minutes 3.981 seconds)