Peak integration¶
This tutorial shows how to find peak maxima and determine peak areas with spectrochempy. As prerequisite, the user is expected to have read the Import, Import IR, slicing and baseline correction tutorials.
First lets import the SpectroChemPy API
[1]:
import spectrochempy as scp
|
|
SpectroChemPy's API - v.0.6.9.dev9 © Copyright 2014-2024 - A.Travert & C.Fernandez @ LCS |
Now import some 2D data into a NDDataset object
[2]:
ds = scp.read_omnic("irdata/nh4y-activation.spg")
ds
[2]:
| name | nh4y-activation |
| author | runner@fv-az1106-694 |
| created | 2024-04-27 03:04:48+02:00 |
| description | Omnic title: NH4Y-activation.SPG Omnic filename: /home/runner/.spectrochempy/testdata/irdata/nh4y-activation.spg |
| history | 2024-04-27 03:04:48+02:00> Imported from spg file /home/runner/.spectrochempy/testdata/irdata/nh4y-activation.spg. 2024-04-27 03:04:48+02:00> Sorted by date |
| DATA | |
| title | absorbance |
| values | [[ 2.057 2.061 ... 2.013 2.012] [ 2.033 2.037 ... 1.913 1.911] ... [ 1.794 1.791 ... 1.198 1.198] [ 1.816 1.815 ... 1.24 1.238]] a.u. |
| shape | (y:55, x:5549) |
| DIMENSION `x` | |
| size | 5549 |
| title | wavenumbers |
| coordinates | [ 6000 5999 ... 650.9 649.9] cm⁻¹ |
| DIMENSION `y` | |
| size | 55 |
| title | acquisition timestamp (GMT) |
| coordinates | [1.468e+09 1.468e+09 ... 1.468e+09 1.468e+09] s |
| labels | [[ 2016-07-06 19:03:14+00:00 2016-07-06 19:13:14+00:00 ... 2016-07-07 04:03:17+00:00 2016-07-07 04:13:17+00:00] [ vz0466.spa, Wed Jul 06 21:00:38 2016 (GMT+02:00) vz0467.spa, Wed Jul 06 21:10:38 2016 (GMT+02:00) ... vz0520.spa, Thu Jul 07 06:00:41 2016 (GMT+02:00) vz0521.spa, Thu Jul 07 06:10:41 2016 (GMT+02:00)]] |
It’s a series of 55 spectra.
For the demonstration select only the first 20 on a limited region from 1250 to 1800 cm\(^{-1}\) (Do not forget to use floating numbers for slicing)
[3]:
X = ds[:20, 1250.0:1800.0]
We can also eventually remove offset on the acquisition time dimension (y)
[4]:
X.y -= X.y[0]
X.y.ito("min")
X.y.title = "acquisition time"
We set some plotting preferences and then plot the raw data
[5]:
prefs = X.preferences
prefs.figure.figsize = (6, 3)
prefs.colormap = "Dark2"
prefs.colorbar = True
X.plot()
[5]:
<_Axes: xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
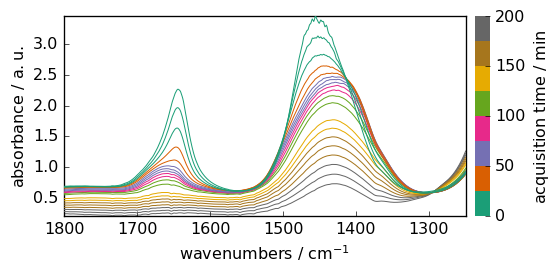
Now we can perform some baseline correction
[6]:
blc = scp.Baseline()
blc.ranges = (
[1740.0, 1800.0],
[1550.0, 1570.0],
[1250.0, 1300.0],
) # define 3 regions where we want the baseline to reach zero.
blc.model = "polynomial"
blc.order = 3
blc.fit(X) # fit the baseline
Xcorr = blc.corrected # get the corrected dataset
Xcorr.plot()
[6]:
<_Axes: xlabel='wavenumbers $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
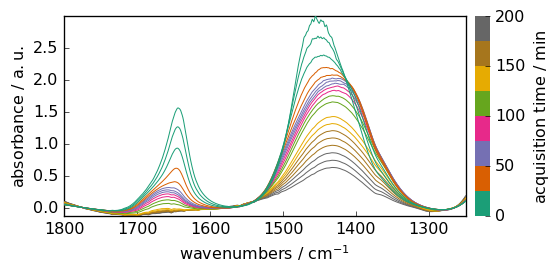
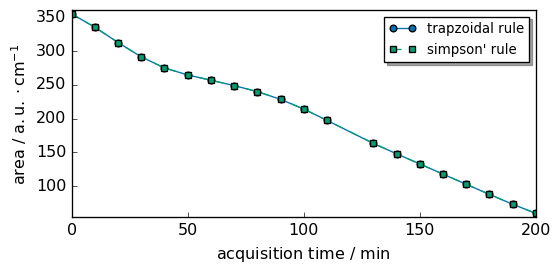
To integrate each row on the full range, we can use the sum or trapz method of a NDDataset.
[7]:
inttrapz = Xcorr.trapezoid(dim="x")
intsimps = Xcorr.simpson(dim="x")
WARNING | (DeprecationWarning) 'scipy.integrate.trapz' is deprecated in favour of 'scipy.integrate.trapezoid' and will be removed in SciPy 1.14.0
WARNING | (DeprecationWarning) 'scipy.integrate.simps' is deprecated in favour of 'scipy.integrate.simpson' and will be removed in SciPy 1.14.0
As you can see both method give almost the same results in this case
[8]:
scp.plot_multiple(
method="scatter",
ms=5,
datasets=[inttrapz, intsimps],
labels=["trapzoidal rule", "simpson' rule"],
legend="best",
)
[8]:
<_Axes: xlabel='acquisition time $\\mathrm{/\\ \\mathrm{min}}$', ylabel='area $\\mathrm{/\\ \\mathrm{a.u.} \\cdot \\mathrm{cm}^{-1}}$'>
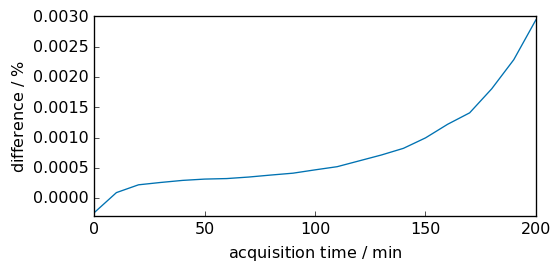
The difference between the trapezoidal and simpson integration methods is visualized below. In this case they are extremely close.
[9]:
diff = (inttrapz - intsimps) * 100.0 / intsimps
diff.title = "difference"
diff.units = "percent"
diff.plot(scatter=True, ms=5)
[9]:
<_Axes: xlabel='acquisition time $\\mathrm{/\\ \\mathrm{min}}$', ylabel='difference $\\mathrm{/\\ \\mathrm{\\%}}$'>