Mathematical operations¶
[1]:
import numpy as np
import spectrochempy as scp
from spectrochempy import MASKED, DimensionalityError, error_
|
|
SpectroChemPy's API - v.0.6.9.dev9 © Copyright 2014-2024 - A.Travert & C.Fernandez @ LCS |
Ufuncs (Universal Numpy’s functions)¶
A universal function (or ufunc in short) is a function that operates on numpy arrays in an element-by-element fashion, supporting array broadcasting, type casting, and several other standard features. That is, a ufunc is a “vectorized” wrapper for a function that takes a fixed number of specific inputs and produces a fixed number of specific outputs.
For instance, in numpy to calculate the square root of each element of a given nd-array, we can write something like this using the np.sqrt functions :
[2]:
x = np.array([1.0, 2.0, 3.0, 4.0, 6.0])
np.sqrt(x)
[2]:
array([ 1, 1.414, 1.732, 2, 2.449])
As seen above, np.sqrt(x) return a numpy array.
The interesting thing, it that ufunc’s can also work with NDDataset .
[3]:
dx = scp.NDDataset(x)
np.sqrt(dx)
[3]:
| name | NDDataset_4dcb3d58 |
| author | runner@fv-az1501-19 |
| created | 2024-04-28 03:11:55+02:00 |
| history | 2024-04-28 03:11:55+02:00> Ufunc sqrt applied. |
| DATA | |
| title | sqrt( |
| values | [ 1 1.414 1.732 2 2.449] |
| size | 5 |
List of UFuncs working on NDDataset:¶
Functions affecting magnitudes of the number but keeping units¶
negative(x, **kwargs): Numerical negative, element-wise.
absolute(x, **kwargs): Calculate the absolute value, element-wise. Alias: abs
fabs(x, **kwargs): Calculate the absolute value, element-wise. Complex values are not handled, use absolute to find the absolute values of complex data.
conj(x, **kwargs): Return the complex conjugate, element-wise.
rint(x, **kwargs) :Round to the nearest integer, element-wise.
floor(x, **kwargs): Return the floor of the input, element-wise.
ceil(x, **kwargs): Return the ceiling of the input, element-wise.
trunc(x, **kwargs): Return the truncated value of the input, element-wise.
Functions affecting magnitudes of the number but also units¶
sqrt(x, **kwargs): Return the non-negative square-root of an array, element-wise.
square(x, **kwargs): Return the element-wise square of the input.
cbrt(x, **kwargs): Return the cube-root of an array, element-wise.
reciprocal(x, **kwargs): Return the reciprocal of the argument, element-wise.
Functions that require no units or dimensionless units for inputs. Returns dimensionless objects.¶
exp(x, **kwargs): Calculate the exponential of all elements in the input array.
exp2(x, **kwargs): Calculate 2**p for all p in the input array.
expm1(x, **kwargs): Calculate
exp(x) - 1for all elements in the array.log(x, **kwargs): Natural logarithm, element-wise.
log2(x, **kwargs): Base-2 logarithm of x.
log10(x, **kwargs): Return the base 10 logarithm of the input array, element-wise.
log1p(x, **kwargs): Return
log(x + 1), element-wise.
Functions that return numpy arrays (Work only for NDDataset)¶
sign(x): Returns an element-wise indication of the sign of a number.
logical_not(x): Compute the truth value of NOT x element-wise.
isfinite(x): Test element-wise for finiteness.
isinf(x): Test element-wise for positive or negative infinity.
isnan(x): Test element-wise for
NaNand return result as a boolean array.signbit(x): Returns element-wise
Truewhere signbit is set.
Trigonometric functions. Require unitless data or radian units.¶
sin(x, **kwargs): Trigonometric sine, element-wise.
cos(x, **kwargs): Trigonometric cosine element-wise.
tan(x, **kwargs): Compute tangent element-wise.
arcsin(x, **kwargs): Inverse sine, element-wise.
arccos(x, **kwargs): Trigonometric inverse cosine, element-wise.
arctan(x, **kwargs): Trigonometric inverse tangent, element-wise.
Hyperbolic functions¶
sinh(x, **kwargs): Hyperbolic sine, element-wise.
cosh(x, **kwargs): Hyperbolic cosine, element-wise.
tanh(x, **kwargs): Compute hyperbolic tangent element-wise.
arcsinh(x, **kwargs): Inverse hyperbolic sine element-wise.
arccosh(x, **kwargs): Inverse hyperbolic cosine, element-wise.
arctanh(x, **kwargs): Inverse hyperbolic tangent element-wise.
Unit conversions¶
Binary Ufuncs¶
add(x1, x2, **kwargs): Add arguments element-wise.
subtract(x1, x2, **kwargs): Subtract arguments, element-wise.
multiply(x1, x2, **kwargs): Multiply arguments element-wise.
divide or true_divide(x1, x2, **kwargs): Returns a true division of the inputs, element-wise.
floor_divide(x1, x2, **kwargs): Return the largest integer smaller or equal to the division of the inputs.
mod or remainder(x1, x2,**kwargs): Return element-wise remainder of division.
fmod(x1, x2, **kwargs): Return the element-wise remainder of division.
Usage¶
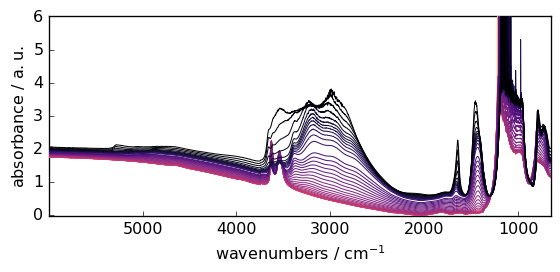
To demonstrate the use of mathematical operations on spectrochempy object, we will first load an experimental 2D dataset.
[4]:
d2D = scp.read_omnic("irdata/nh4y-activation.spg")
prefs = d2D.preferences
prefs.colormap = "magma"
prefs.colorbar = False
prefs.figure.figsize = (6, 3)
_ = d2D.plot()
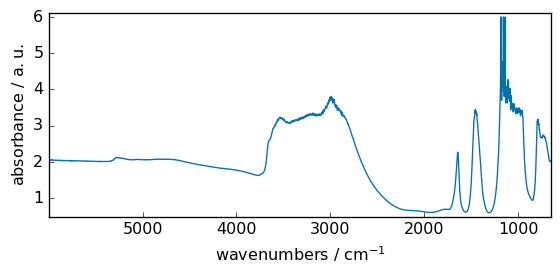
Let’s select only the first row of the 2D dataset ( the squeeze method is used to remove the residual size 1 dimension). In addition, we mask the saturated region.
[5]:
dataset = d2D[0].squeeze()
_ = dataset.plot()
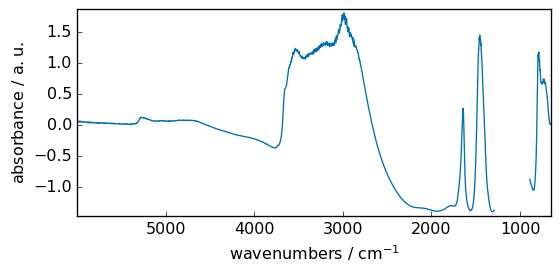
This dataset will be artificially modified already using some mathematical operation (subtraction with a scalar) to present negative values, and we will also mask some data
[6]:
dataset -= 2.0 # add an offset to make that some of the values become negative
dataset[1290.0:890.0] = scp.MASKED # additionally we mask some data
_ = dataset.plot()
Unary functions¶
Functions affecting magnitudes of the number but keeping units¶
negative¶
Numerical negative, element-wise, keep units
[7]:
out = np.negative(dataset) # the same results is obtained using out=-dataset
_ = out.plot(figsize=(6, 2.5), show_mask=True)
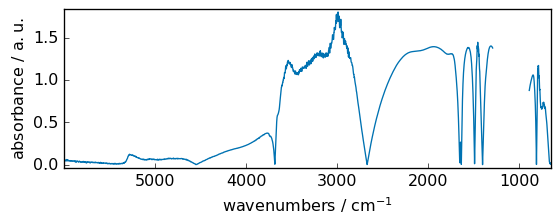
abs¶
absolute (alias of abs)¶
fabs (absolute for float arrays)¶
Numerical absolute value element-wise, element-wise, keep units
[8]:
out = np.abs(dataset)
_ = out.plot(figsize=(6, 2.5))
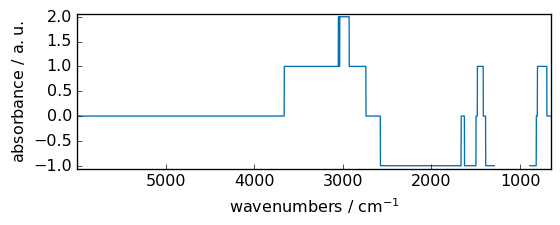
rint¶
Round elements of the array to the nearest integer, element-wise, keep units
[9]:
out = np.rint(dataset)
_ = out.plot(figsize=(6, 2.5)) # not that title is not modified for this ufunc
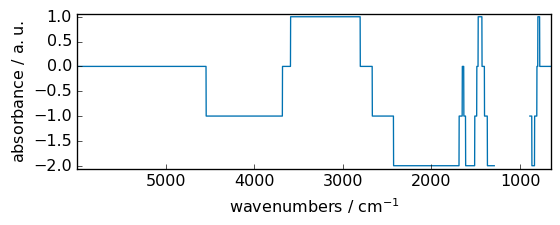
floor¶
Return the floor of the input, element-wise.
[10]:
out = np.floor(dataset)
_ = out.plot(figsize=(6, 2.5))
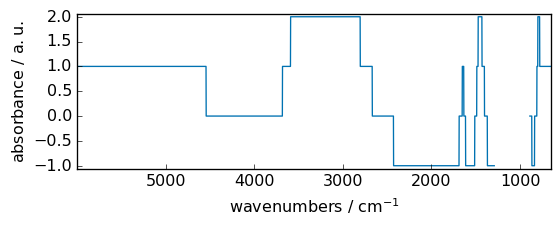
ceil¶
Return the ceiling of the input, element-wise.
[11]:
out = np.ceil(dataset)
_ = out.plot(figsize=(6, 2.5))
trunc¶
Return the truncated value of the input, element-wise.
[12]:
out = np.trunc(dataset)
_ = out.plot(figsize=(6, 2.5))
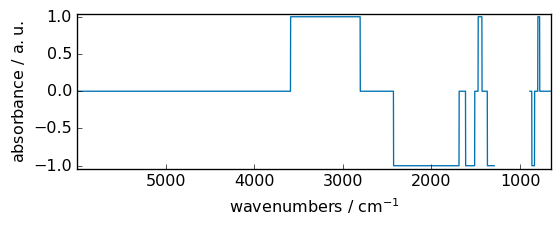
Functions affecting magnitudes of the number but also units¶
sqrt¶
Return the non-negative square-root of an array, element-wise.
[13]:
out = np.sqrt(
dataset
) # as they are some negative elements, return dataset has complex dtype.
_ = out.plot_1D(show_complex=True, figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
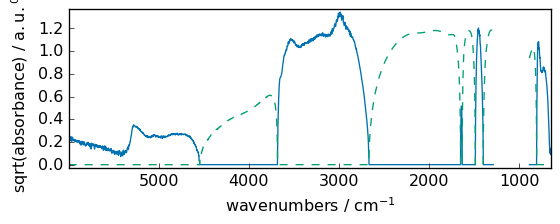
square¶
Return the element-wise square of the input.
[14]:
out = np.square(dataset)
_ = out.plot(figsize=(6, 2.5))
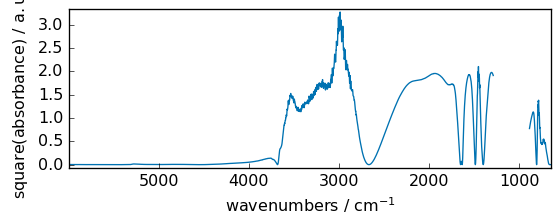
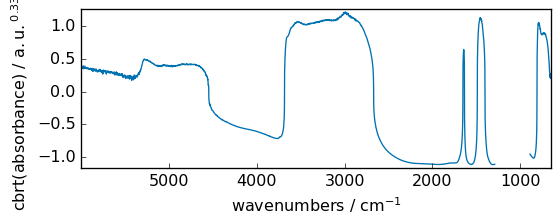
cbrt¶
Return the cube-root of an array, element-wise.
[15]:
out = np.cbrt(dataset)
_ = out.plot(figsize=(6, 2.5))
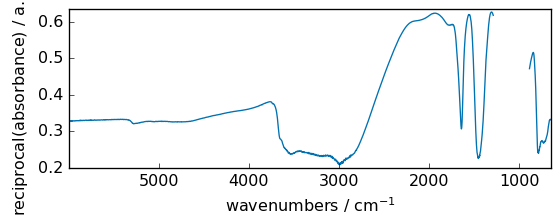
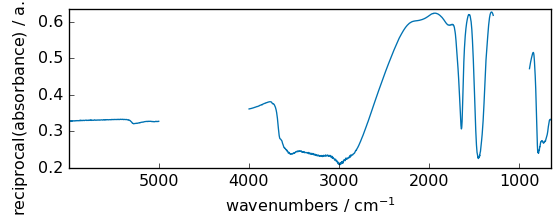
reciprocal¶
Return the reciprocal of the argument, element-wise.
[16]:
out = np.reciprocal(dataset + 3.0)
_ = out.plot(figsize=(6, 2.5))
Functions that require no units or dimensionless units for inputs. Returns dimensionless objects.¶
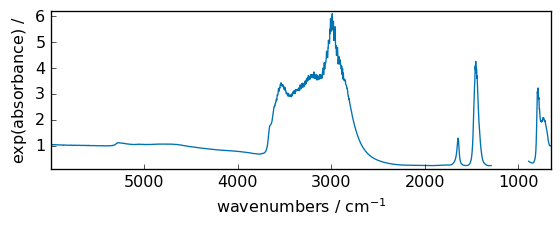
exp¶
Exponential of all elements in the input array, element-wise
[17]:
out = np.exp(dataset)
_ = out.plot(figsize=(6, 2.5))
Obviously numpy exponential functions applies only to dimensionless array. Else an error is generated.
[18]:
x = scp.NDDataset(np.arange(5), units="m")
try:
np.exp(x) # A dimensionality error will be generated
except DimensionalityError as e:
error_(DimensionalityError, e)
ERROR | DimensionalityError: Cannot convert from 'm' to ''
Function `exp` requires DIMENSIONLESS input
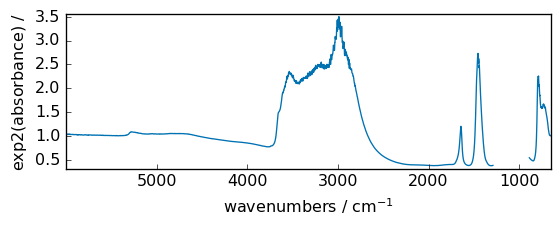
exp2¶
Calculate 2**p for all p in the input array.
[19]:
out = np.exp2(dataset)
_ = out.plot(figsize=(6, 2.5))
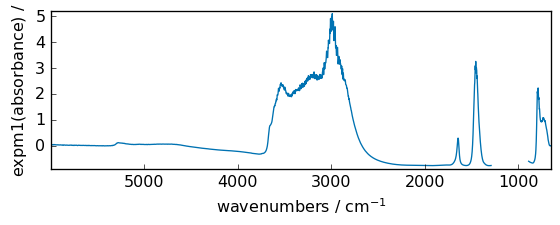
expm1¶
Calculate exp(x) - 1 for all elements in the array.
[20]:
out = np.expm1(dataset)
_ = out.plot(figsize=(6, 2.5))
log¶
Natural logarithm, element-wise.
This doesn’t generate un error for negative numbrs, but the output is masked for those values
[21]:
out = np.log(dataset)
ax = out.plot(figsize=(6, 2.5), show_mask=True)
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
[22]:
out = np.log(dataset - dataset.min())
_ = out.plot(figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
log2¶
Base-2 logarithm of x.
[23]:
out = np.log2(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
log10¶
Return the base 10 logarithm of the input array, element-wise.
[24]:
out = np.log10(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
log1p¶
Return log(x + 1) , element-wise.
[25]:
out = np.log1p(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
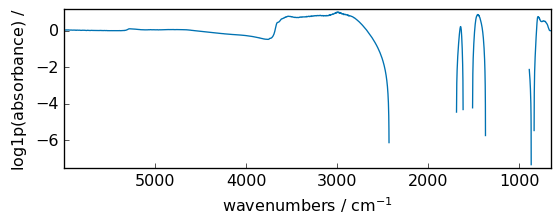
Functions that return numpy arrays (Work only for NDDataset)¶
sign¶
Returns an element-wise indication of the sign of a number. Returned object is a ndarray
[26]:
np.sign(dataset)
[26]:
masked_array(data=[ 1, 1, ..., 1, 1],
mask=[ False, False, ..., False, False],
fill_value=1e+20)
[27]:
np.logical_not(dataset < 0)
[27]:
masked_array(data=[ 1, 1, ..., 1, 1],
mask=[ False, False, ..., False, False],
fill_value=True)
isfinite¶
Test element-wise for finiteness.
[28]:
np.isfinite(dataset)
[28]:
masked_array(data=[ 1, 1, ..., 1, 1],
mask=[ False, False, ..., False, False],
fill_value=True)
isinf¶
Test element-wise for positive or negative infinity.
[29]:
np.isinf(dataset)
[29]:
masked_array(data=[ 0, 0, ..., 0, 0],
mask=[ False, False, ..., False, False],
fill_value=True)
isnan¶
Test element-wise for NaN and return result as a boolean array.
[30]:
np.isnan(dataset)
[30]:
masked_array(data=[ 0, 0, ..., 0, 0],
mask=[ False, False, ..., False, False],
fill_value=True)
signbit¶
Returns element-wise True where signbit is set.
[31]:
np.signbit(dataset)
[31]:
masked_array(data=[ 0, 0, ..., 0, 0],
mask=[ False, False, ..., False, False],
fill_value=True)
Trigonometric functions. Require dimensionless/unitless dataset or radians.¶
In the below examples, unit of data in dataset is absorbance (then dimensionless)
sin¶
Trigonometric sine, element-wise.
[32]:
out = np.sin(dataset)
_ = out.plot(figsize=(6, 2.5))
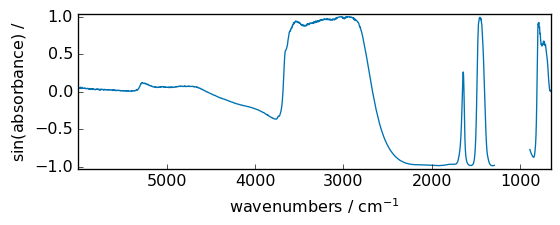
cos¶
Trigonometric cosine element-wise.
[33]:
out = np.cos(dataset)
_ = out.plot(figsize=(6, 2.5))
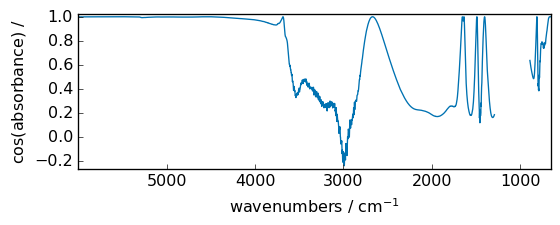
tan¶
Compute tangent element-wise.
[34]:
out = np.tan(dataset / np.max(dataset))
_ = out.plot(figsize=(6, 2.5))
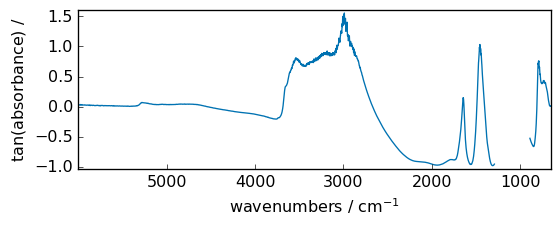
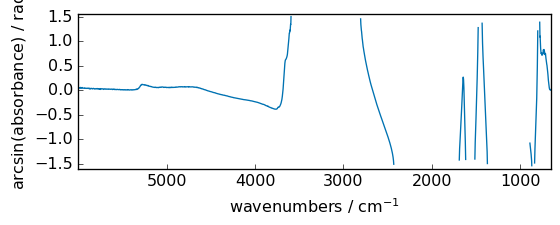
arcsin¶
Inverse sine, element-wise.
[35]:
out = np.arcsin(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (RuntimeWarning) invalid value encountered in arcsin
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
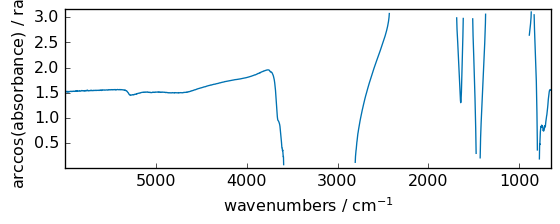
arccos¶
Trigonometric inverse cosine, element-wise.
[36]:
out = np.arccos(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (RuntimeWarning) invalid value encountered in arccos
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
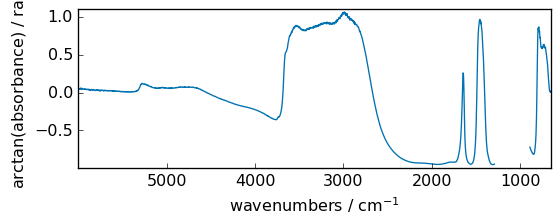
arctan¶
Trigonometric inverse tangent, element-wise.
[37]:
out = np.arctan(dataset)
_ = out.plot(figsize=(6, 2.5))
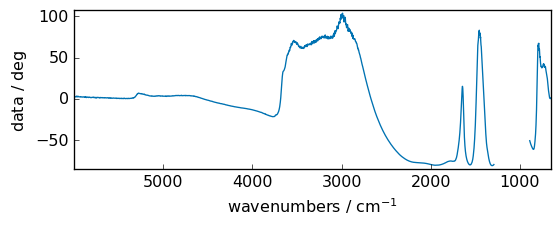
Angle units conversion¶
rad2deg¶
Convert angles from radians to degrees (warning: unitless or dimensionless are assumed to be radians, so no error will be issued).
for instance, if we take the z axis (the data magnitude) in the figure above, it’s expressed in radians. We can change to degrees easily.
[38]:
out = np.rad2deg(dataset)
out.title = "data" # just to avoid a too long title
_ = out.plot(figsize=(6, 2.5))
deg2rad¶
Convert angles from degrees to radians.
[39]:
out = np.deg2rad(out)
out.title = "data"
_ = out.plot(figsize=(6, 2.5))
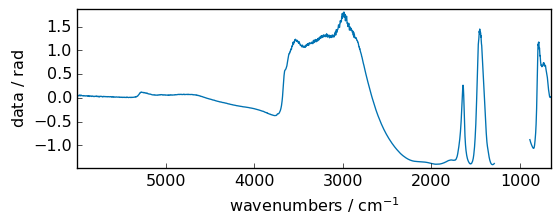
Hyperbolic functions¶
sinh¶
Hyperbolic sine, element-wise.
[40]:
out = np.sinh(dataset)
_ = out.plot(figsize=(6, 2.5))
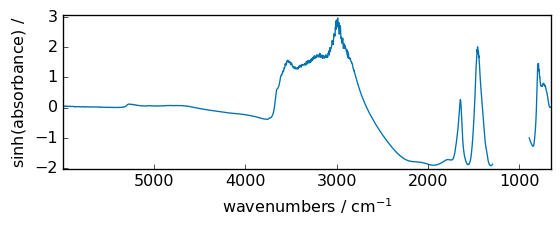
cosh¶
Hyperbolic cosine, element-wise.
[41]:
out = np.cosh(dataset)
_ = out.plot(figsize=(6, 2.5))
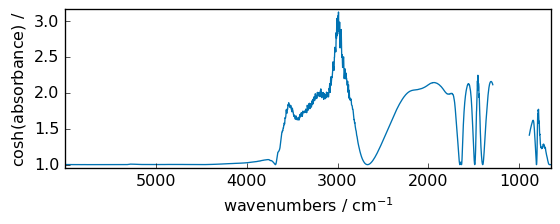
tanh¶
Compute hyperbolic tangent element-wise.
[42]:
out = np.tanh(dataset)
_ = out.plot(figsize=(6, 2.5))
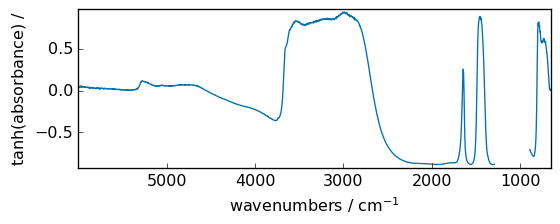
arcsinh¶
Inverse hyperbolic sine element-wise.
[43]:
out = np.arcsinh(dataset)
_ = out.plot(figsize=(6, 2.5))
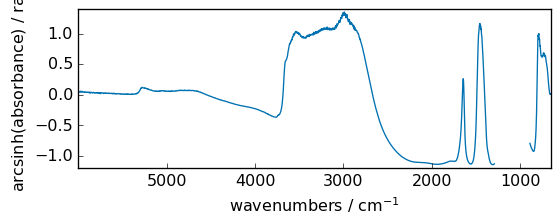
arccosh¶
Inverse hyperbolic cosine, element-wise.
[44]:
out = np.arccosh(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
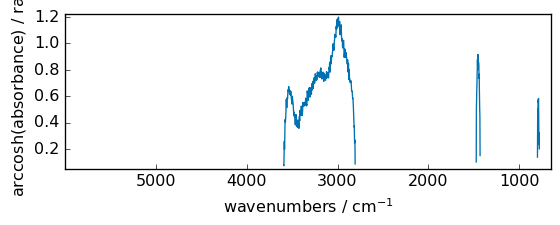
arctanh¶
Inverse hyperbolic tangent element-wise.
[45]:
out = np.arctanh(dataset)
_ = out.plot(figsize=(6, 2.5))
WARNING | (RuntimeWarning) invalid value encountered in arctanh
WARNING | (UserWarning) Given trait value dtype "float64" does not match required type "float64". A coerced copy has been created.
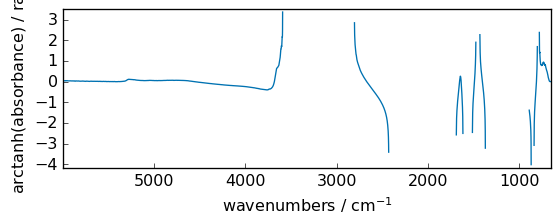
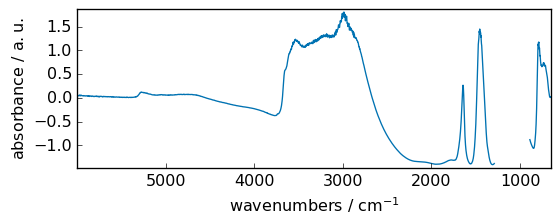
Binary functions¶
[46]:
dataset2 = np.reciprocal(dataset + 3) # create a second dataset
dataset2[5000.0:4000.0] = MASKED
_ = dataset.plot(figsize=(6, 2.5))
_ = dataset2.plot(figsize=(6, 2.5))
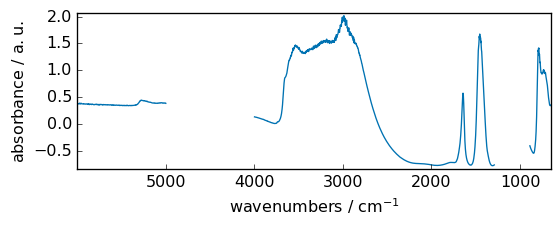
Arithmetic¶
add¶
Add arguments element-wise.
[47]:
out = np.add(dataset, dataset2)
_ = out.plot(figsize=(6, 2.5))
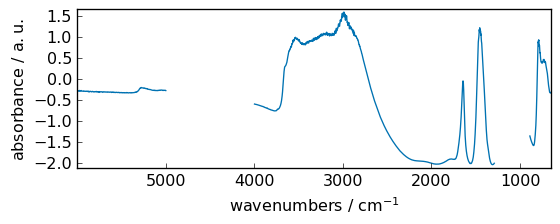
subtract¶
Subtract arguments, element-wise.
[48]:
out = np.subtract(dataset, dataset2)
_ = out.plot(figsize=(6, 2.5))
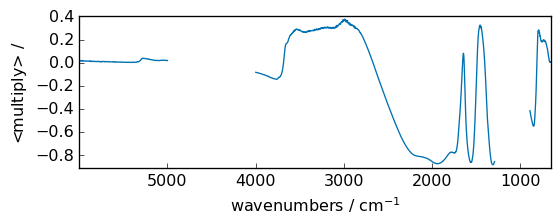
multiply¶
Multiply arguments element-wise.
[49]:
out = np.multiply(dataset, dataset2)
_ = out.plot(figsize=(6, 2.5))
divide¶
or ##### true_divide Returns a true division of the inputs, element-wise.
[50]:
out = np.divide(dataset, dataset2)
_ = out.plot(figsize=(6, 2.5))
floor_divide¶
Return the largest integer smaller or equal to the division of the inputs.
[51]:
out = np.floor_divide(dataset, dataset2)
_ = out.plot(figsize=(6, 2.5))
Complex or hypercomplex NDDatasets¶
NDDataset objects with complex data are handled differently than in numpy.ndarray .
Instead, complex data are stored by interlacing the real and imaginary part. This allows the definition of data that can be complex in several axis, and e .g., allows 2D-hypercomplex array that can be transposed (useful for NMR data).
[52]:
da = scp.NDDataset(
[
[1.0 + 2.0j, 2.0 + 0j],
[1.3 + 2.0j, 2.0 + 0.5j],
[1.0 + 4.2j, 2.0 + 3j],
[5.0 + 4.2j, 2.0 + 3j],
]
)
da
[52]:
| name | NDDataset_532fc138 |
| author | runner@fv-az1501-19 |
| created | 2024-04-28 03:12:04+02:00 |
| DATA | |
| title | |
| values | R[[ 1 2] [ 1.3 2] [ 1 2] [ 5 2]] I[[ 2 0] [ 2 0.5] [ 4.2 3] [ 4.2 3]] |
| shape | (y:4, x:2(complex)) |
A dataset of type float can be transformed into a complex dataset (using two consecutive rows to create a complex row)
[53]:
da = scp.NDDataset(np.arange(40).reshape(10, 4))
da
[53]:
| name | NDDataset_53310afc |
| author | runner@fv-az1501-19 |
| created | 2024-04-28 03:12:04+02:00 |
| DATA | |
| title | |
| values | [[ 0 1 2 3] [ 4 5 6 7] ... [ 32 33 34 35] [ 36 37 38 39]] |
| shape | (y:10, x:4) |
[54]:
dac = da.set_complex()
dac
[54]:
| name | NDDataset_5331ffac |
| author | runner@fv-az1501-19 |
| created | 2024-04-28 03:12:04+02:00 |
| DATA | |
| title | |
| values | R[[ 0 2] [ 4 6] ... [ 32 34] [ 36 38]] I[[ 1 3] [ 5 7] ... [ 33 35] [ 37 39]] |
| shape | (y:10, x:2(complex)) |
Note the xdimension size is divided by a factor of two
A dataset which is complex in two dimensions is called hypercomplex (it’s datatype in SpectroChemPy is set to quaternion).
[55]:
daq = da.set_quaternion() # equivalently one can use the set_hypercomplex method
daq
[55]:
| name | NDDataset_53334e84 |
| author | runner@fv-az1501-19 |
| created | 2024-04-28 03:12:04+02:00 |
| DATA | |
| title | |
| values | RR[[ 0 2] [ 8 10] ... [ 24 26] [ 32 34]] RI[[ 1 3] [ 9 11] ... [ 25 27] [ 33 35]] IR[[ 4 6] [ 12 14] ... [ 28 30] [ 36 38]] II[[ 5 7] [ 13 15] ... [ 29 31] [ 37 39]] |
| shape | (y:5(complex), x:2(complex)) |
[56]:
daq.dtype
[56]:
dtype(quaternion)