Time domain baseline correction (NMR)¶
Here we show how to baseline correct dataset in the time domain before applying FFT.
The example spectra were downloaded from this page where you can find some explanations on this kind of process.
[1]:
import spectrochempy as scp
|
|
SpectroChemPy's API - v.0.6.9.dev9 © Copyright 2014-2024 - A.Travert & C.Fernandez @ LCS |
[2]:
path = scp.preferences.datadir / "nmrdata" / "bruker" / "tests" / "nmr" / "h3po4"
fid = scp.read_topspin(path, expno=4)
prefs = fid.preferences
prefs.figure.figsize = (7, 3)
_ = fid.plot(show_complex=True)
WARNING | (UserWarning) Error reading the pulse program
[3]:
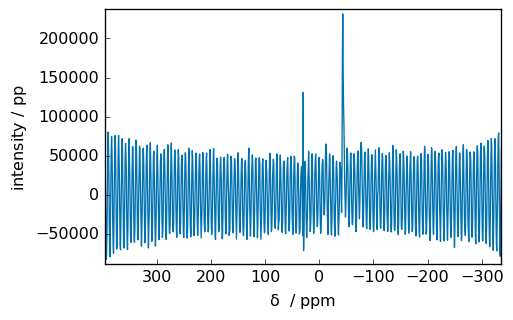
spec = scp.fft(fid)
_ = spec.plot(xlim=(5, -5))
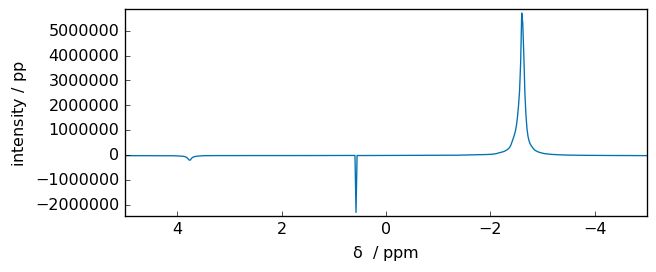
We can see that in the middle of the spectrum there are an artifact (a transmitter spike) due to different DC offset between imaginary.
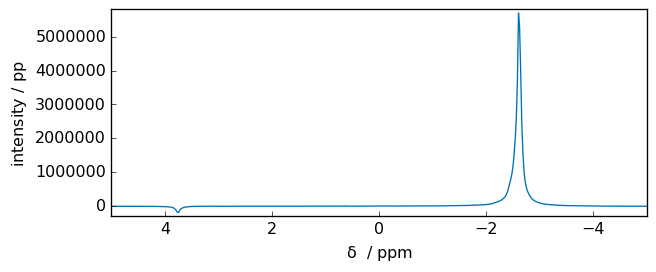
In SpectroChemPy, for now, we provide a simple kind of dc correction using the dccommand.
[4]:
dc_corrected_fid = fid.dc()
spec = scp.fft(dc_corrected_fid)
_ = spec.plot(xlim=(5, -5))
[5]:
path = scp.preferences.datadir / "nmrdata" / "bruker" / "tests" / "nmr" / "cadmium"
fid2 = scp.read_topspin(path, expno=100)
_ = fid2.plot(show_complex=True)
WARNING | (UserWarning) Error reading the pulse program
WARNING | (UserWarning) (956,)cannot be shaped into(1024,)
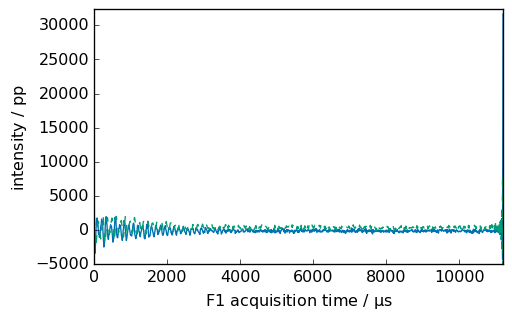
[6]:
spec2 = scp.fft(fid2)
_ = spec2.plot()
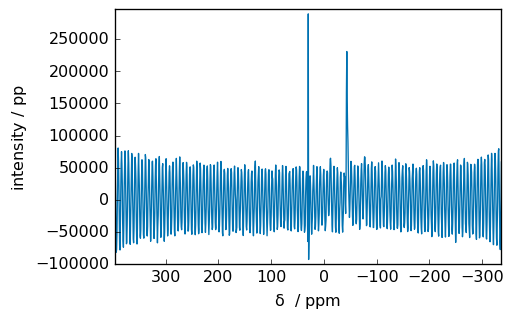
[7]:
dc_corrected_fid2 = fid2.dc()
spec2 = scp.fft(dc_corrected_fid2)
_ = spec2.plot()