Note
Go to the end to download the full example code
NDDataset creation and plotting example¶
In this example, we create a 3D NDDataset from scratch, and then we plot one section (a 2D plane)
Creation¶
Now we will create a 3D NDDataset from scratch
Data¶
here we make use of numpy array functions to create the data for coordinates axis and the array of data
import numpy as np
As usual, we start by loading the spectrochempy library
import spectrochempy as scp
c0 = np.linspace(200.0, 300.0, 3)
c1 = np.linspace(0.0, 60.0, 100)
c2 = np.linspace(4000.0, 1000.0, 100)
nd_data = np.array(
[
np.array([np.sin(2.0 * np.pi * c2 / 4000.0) * np.exp(-y / 60) for y in c1]) * t
for t in c0
]
)
Coordinates¶
The Coord object allow making an array of coordinates
with additional metadata such as units, labels, title, etc
Labels can be useful for instance for indexing
Coord: [float64] K (size: 1)
nd-Dataset¶
The NDDataset object allow making the array of data with units, etc…
mydataset = scp.NDDataset(
nd_data, coordset=[coord0, coord1, coord2], title="Absorbance", units="absorbance"
)
mydataset.description = """Dataset example created for this tutorial.
It's a 3-D dataset (with dimensionless intensity: absorbance )"""
mydataset.name = "An example from scratch"
mydataset.author = "Blake and Mortimer"
print(mydataset)
NDDataset: [float64] a.u. (shape: (z:3, y:100, x:100))
We want to plot a section of this 3D NDDataset:
NDDataset can be sliced like conventional numpy-array…
or maybe more conveniently in this case, using an axis labels:
To plot a dataset, use the plot command (generic plot).
As the section NDDataset is 2D, a contour plot is displayed by default.
new.plot()
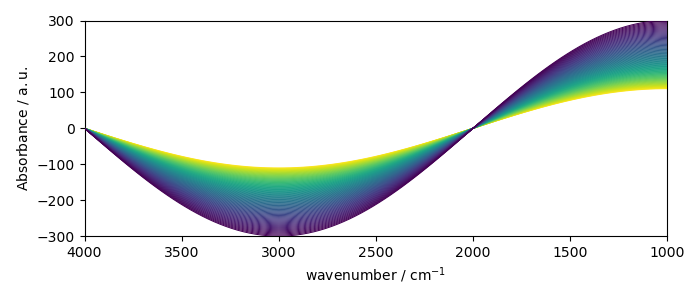
<_Axes: xlabel='wavenumber $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='Absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
But it is possible to display image
sphinx_gallery_thumbnail_number = 2
new.plot(method="image")
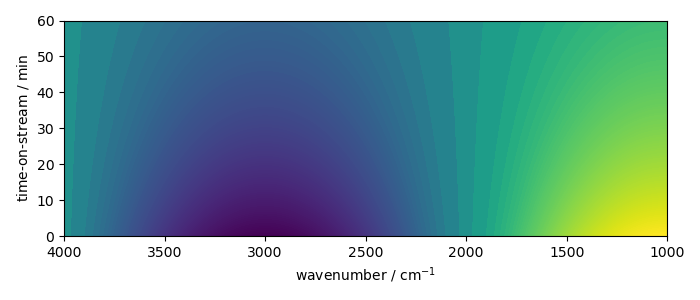
<_Axes: xlabel='wavenumber $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='time-on-stream $\\mathrm{/\\ \\mathrm{min}}$'>
or stacked plot
new.plot(method="stack")
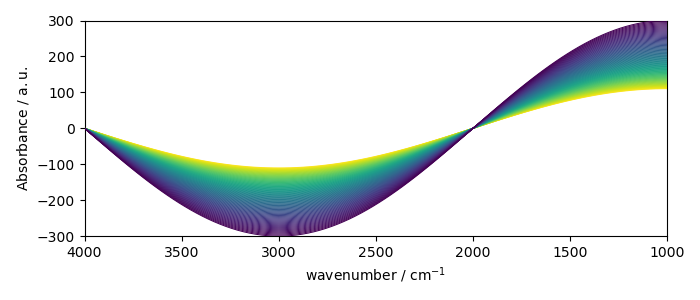
<_Axes: xlabel='wavenumber $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='Absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
Note that the scp allows one to use this syntax too:
<_Axes: xlabel='wavenumber $\\mathrm{/\\ \\mathrm{cm}^{-1}}$', ylabel='Absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script
scp.show()
Total running time of the script: ( 0 minutes 1.604 seconds)