Note
Go to the end to download the full example code
FastICA example¶
Import the spectrochempy API package
import spectrochempy as scp
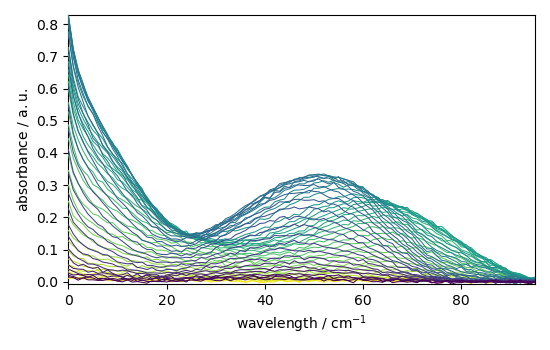
Load, prepare and plot the dataset¶
Here we use a dataset from Jaumot et al. [2005]
Create and fit a FastICA object¶
As argument of the object constructor we define log_level to "INFO" to
obtain verbose output during fit, and we set the number of component to use at 4.
ica = scp.FastICA(n_components=4)
_ = ica.fit(X)
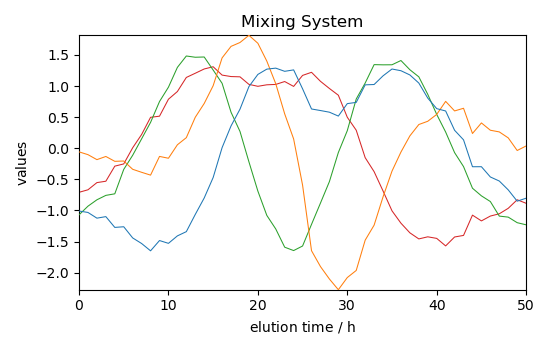
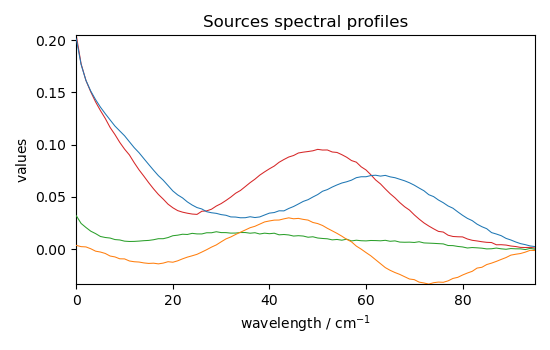
Get the mixing system and source spectral profiles¶
The mixing system \(A\) and the source spectral profiles \(S^T\) can be obtained as follows (the Sklearn equivalents - also valid with Scpy - are indicated as comments
Plot them
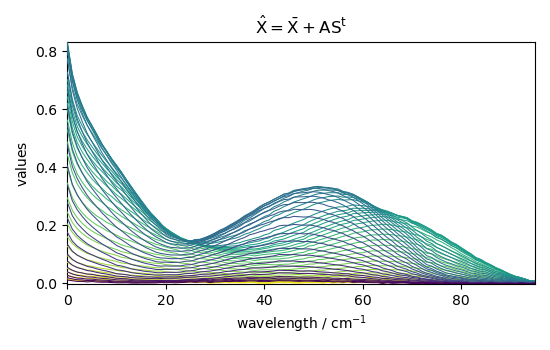
Reconstruct the dataset¶
The dataset can be reconstructed from these matrices and the mean:
Or using the transform() method:
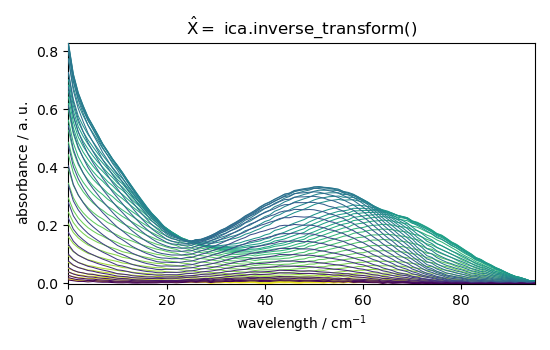
X_hat_b = ica.inverse_transform()
_ = X_hat_b.plot(title="$\hat{X} =$ ica.inverse_transform()")
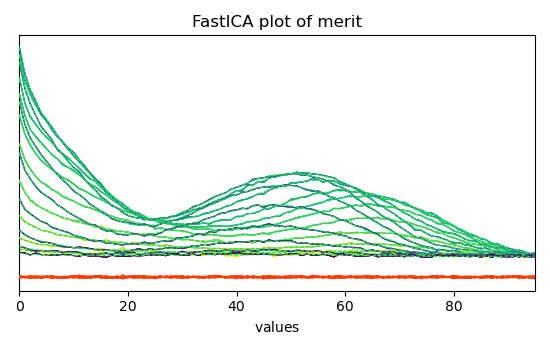
Finally, the quality of the reconstriction can be checked by plotmerit()
_ = ica.plotmerit(nb_traces=15)
This ends the example ! The following line can be uncommented if no plot shows when running the .py script
# scp.show()
Total running time of the script: ( 0 minutes 1.332 seconds)