Note
Go to the end to download the full example code
SIMPLISMA example¶
In this example, we perform the PCA dimensionality reduction of a spectra dataset
Import the spectrochempy API package (and the SIMPLISMA model independently)
import spectrochempy as scp
Load a matlab datasets
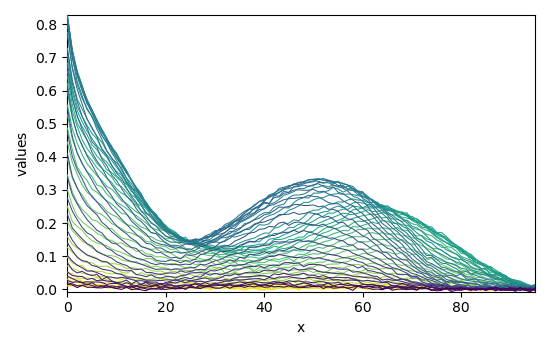
Dataset (Jaumot et al., Chemometr. Intell. Lab. 76 (2005) 101-110)):
cpure (204, 4)
MATRIX (204, 96)
isp_matrix (4, 4)
spure (4, 96)
csel_matrix (51, 4)
m1 (51, 96)
Add some metadata for a nicer display
ds.title = "absorbance"
ds.units = "absorbance"
ds.set_coordset(None, None)
ds.y.title = "elution time"
ds.x.title = "wavelength"
ds.y.units = "hours"
ds.x.units = "nm"
Fit the SIMPLISMA model
print("Fit SIMPLISMA on {}\n".format(ds.name))
simpl = scp.SIMPLISMA(max_components=20, tol=0.2, noise=3, log_level="INFO")
simpl.fit(ds)
Fit SIMPLISMA on m1
/home/runner/micromamba/envs/scpy_docs/lib/python3.10/site-packages/spectrochempy/utils/decorators.py:86: DeprecationWarning: The `max_components` attribute is now deprecated. Use `n_components` instead. `max_components` attribute will be removed in version 0.7.
warn(
/home/runner/micromamba/envs/scpy_docs/lib/python3.10/site-packages/spectrochempy/analysis/decomposition/simplisma.py:190: UserWarning: SIMPLISMA does not handle easily negative values.
warn("SIMPLISMA does not handle easily negative values.")
*** Automatic SIMPL(I)SMA analysis ***
dataset: m1
noise: 3.0 %
tol: 0.2 %
n_components: 20
#iter index_pc coord_pc Std(res) R^2
---------------------------------------------
1 4 4.0 0.0263 0.9755
2 82 82.0 0.0100 0.9964
3 29 29.0 0.0072 0.9981
**** Unexplained variance lower than 'tol' (0.2 %)
**** End of SIMPL(I)SMA analysis.
<spectrochempy.analysis.decomposition.simplisma.SIMPLISMA object at 0x7f1a351a1090>
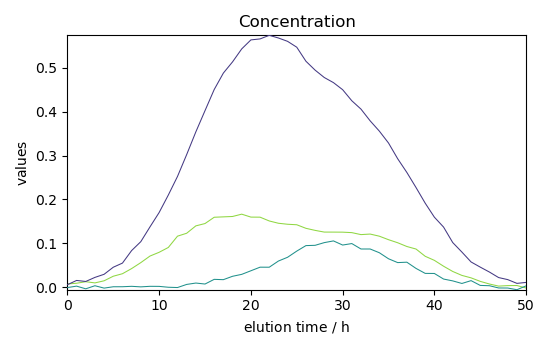
Plot concentration
_ = simpl.C.T.plot(title="Concentration")
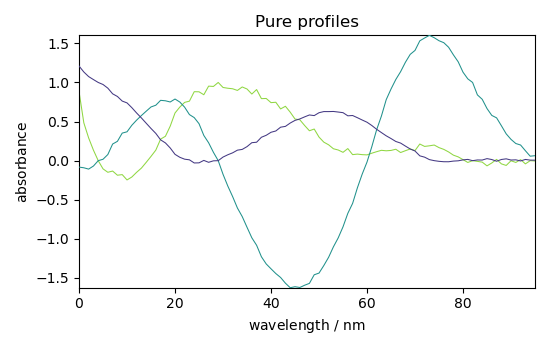
Plot components (St)
_ = simpl.components.plot(title="Pure profiles")
Show the plot of merit after reconstruction oto the original data space
simpl.plotmerit(offset=0, nb_traces=5)
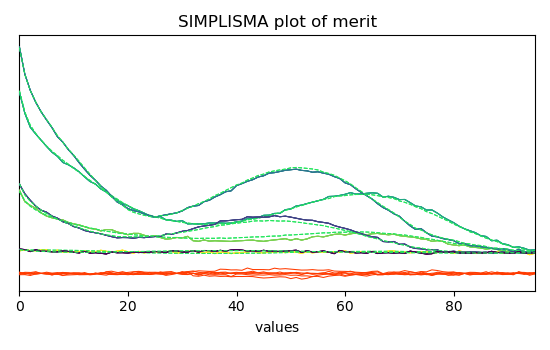
<_Axes: title={'center': 'SIMPLISMA plot of merit'}, xlabel='values $\\mathrm{}$', ylabel='absorbance $\\mathrm{/\\ \\mathrm{a.u.}}$'>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: ( 0 minutes 0.630 seconds)