Note
Go to the end to download the full example code
Exponential window multiplication¶
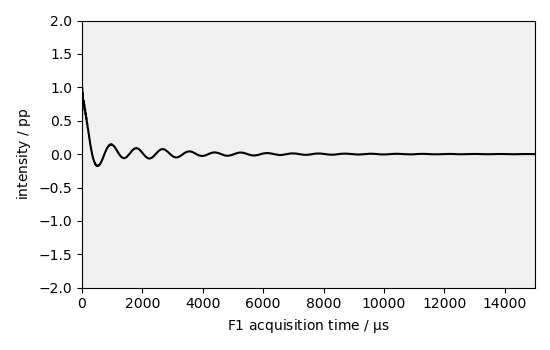
In this example, we perform exponential window multiplication to apodize a NMR signal in the time domain.
import spectrochempy as scp
Hz = scp.ur.Hz
us = scp.ur.us
path = scp.preferences.datadir / "nmrdata" / "bruker" / "tests" / "nmr" / "topspin_1d"
dataset1D = scp.read_topspin(path, expno=1, remove_digital_filter=True)
Normalize the dataset values and reduce the time domain
dataset1D /= dataset1D.real.data.max() # normalize
dataset1D = dataset1D[0.0:15000.0]
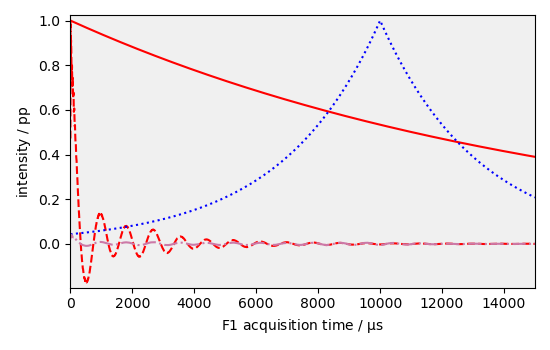
Apply exponential window apodization
new1, curve1 = scp.em(dataset1D.copy(), lb=20 * Hz, retapod=True, inplace=False)
Apply a shifted exponential window apodization default units are HZ for broadening and microseconds for shifting
new2, curve2 = dataset1D.copy().em(
lb=100 * Hz, shifted=10000 * us, retapod=True, inplace=False
)
Plotting
_ = dataset1D.plot(zlim=(-2, 2), color="k")
_ = curve1.plot(color="r")
_ = new1.plot(color="r", clear=False, label=" em = 20 hz")
_ = curve2.plot(color="b", clear=False)
_ = new2.plot(dcolor="b", clear=False, label=" em = 30 HZ, shifted = ")
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: ( 0 minutes 0.504 seconds)