Note
Go to the end to download the full example code
Reading datasets¶
In this example, we show the use of the generic read method to create dataset
either from local or remote files.
First we need to import the spectrochempy API package
import spectrochempy as scp
Import dataset from local files¶
Read a IR data recorded in Omnic format (.spg extension).
We just pass the file name as parameter.
_ = dataset.plot(style="paper")
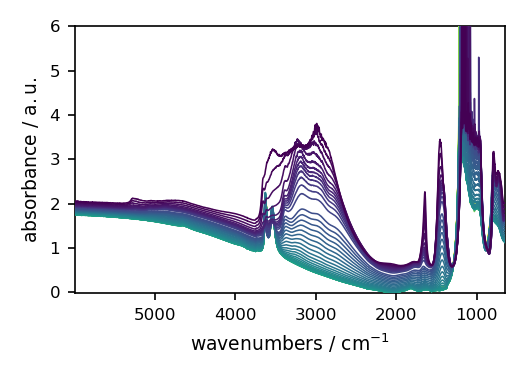
When using read, we can pass filename as a str or a Path object.
Note that is the file is not found in the current working directory, SpectroChemPy
will try to find it in the datadir directory defined in preferences :
PosixPath('/home/runner/.spectrochempy/testdata')
If the supplied argument is a directory, then the whole directory is read at once.
By default, the different files will be merged along the first dimension (y).
However, for this to work, the second dimension (x) must be compatible (same size)
or else a WARNING appears. To avoid the warning and get individual spectra, you can
set merge to False .
dataset_list = scp.read("irdata", merge=False)
dataset_list
[NDDataset: [float64] a.u. (shape: (y:19, x:3112)), NDDataset: [float64] a.u. (shape: (y:55, x:5549)), NDDataset: [float64] unitless (shape: (y:1, x:3736))]
to get full details on the parameters that can be used, look at the API documentation:
spectrochempy.read .
Import dataset from remote files¶
To download and read file from remote server you can use urls.
dataset_list = scp.read("http://www.eigenvector.com/data/Corn/corn.mat")
In this case the matlab data contains 7 arrays that have been automatically
transformed to NDDataset .
for nd in dataset_list:
print(f"{nd.name} : {nd.shape}")
m5nbs : (3, 700)
mp5nbs : (4, 700)
mp6nbs : (4, 700)
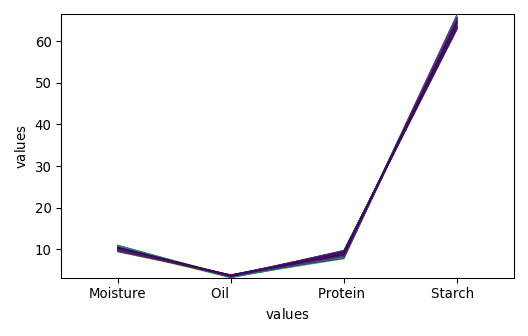
propvals : (80, 4)
m5spec : (80, 700)
mp5spec : (80, 700)
mp6spec : (80, 700)
The eigenvector.com website contains the same data in a
compressed (zipped) format:
corn.mat_.zip .
This can also be used by the read method.
dataset_list = scp.read(
"https://eigenvector.com/wp-content/uploads/2019/06/corn.mat_.zip"
)
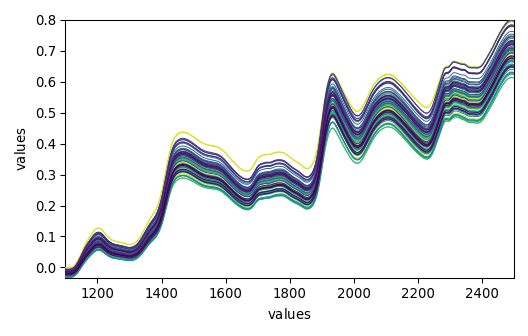
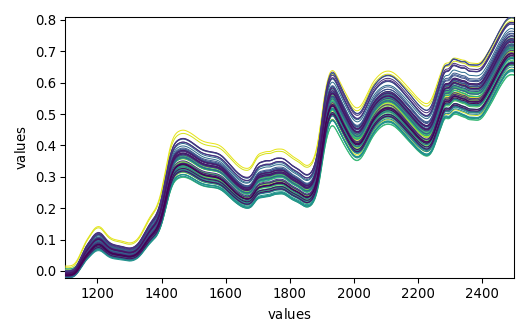
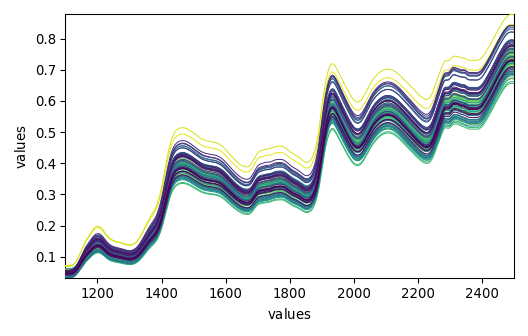
_ = dataset_list[-1].plot()
_ = dataset_list[-2].plot()
_ = dataset_list[-3].plot()
_ = dataset_list[-4].plot()
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: ( 0 minutes 3.825 seconds)