Note
Go to the end to download the full example code.
PCA analysis example
In this example, we perform the PCA dimensionality reduction of a spectra dataset
Import the spectrochempy API package
import spectrochempy as scp
Load a dataset
dataset = scp.read_omnic("irdata/nh4y-activation.spg")[::5]
print(dataset)
dataset.plot()
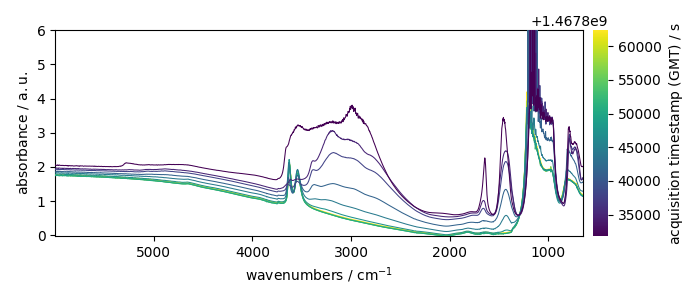
NDDataset: [float64] a.u. (shape: (y:11, x:5549))
Create a PCA object and fit the dataset so that the explained variance is greater or equal to 99.9%
<spectrochempy.analysis.decomposition.pca.PCA object at 0x7f514bd35260>
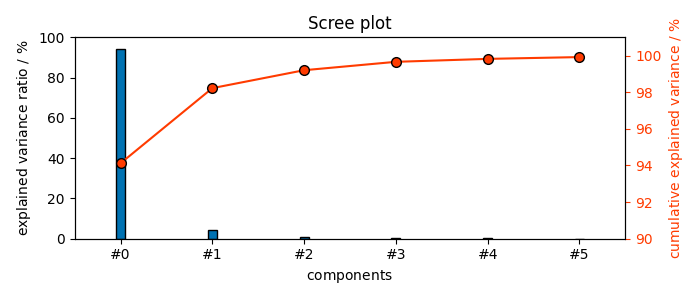
The number of fitted components is given by the n_components attribute (We obtain 23 components)
6
Transform the dataset to a lower dimensionality using all the fitted components
Finally, display the results graphically
# first we can set some preferences for the plot
prefs = scp.preferences
prefs.lines.markersize = 7
# ScreePlot
pca.screeplot()
(<Matplotlib Axes object>, <Axes: xlabel='components $\\mathrm{}$', ylabel='cumulative explained variance $\\mathrm{/\\ \\mathrm{\\%}}$'>)
Score Plot first we can set some preferences for the plot
prefs.lines.markersize = 10
pca.scoreplot(scores, 1, 2)
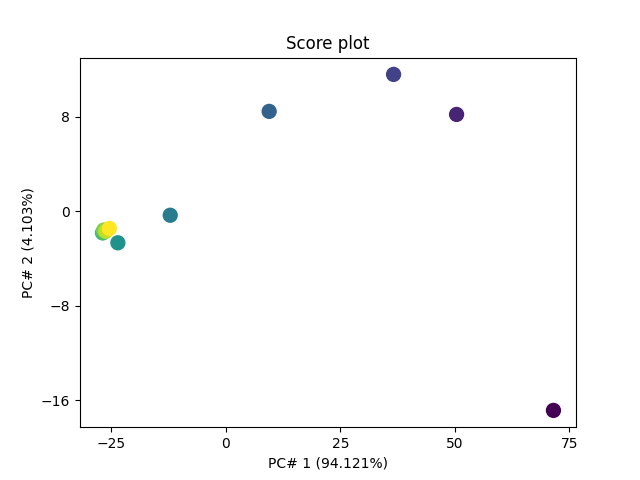
<Axes: title={'center': 'Score plot'}, xlabel='PC# 1 (94.121%)', ylabel='PC# 2 (4.103%)'>
Score Plot for 3 PC’s in 3D
pca.scoreplot(scores, 1, 2, 3)
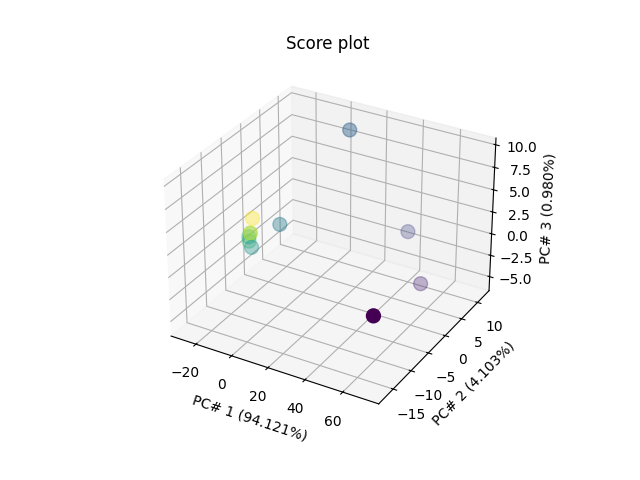
<Axes3D: title={'center': 'Score plot'}, xlabel='PC# 1 (94.121%)', ylabel='PC# 2 (4.103%)', zlabel='PC# 3 (0.980%)'>
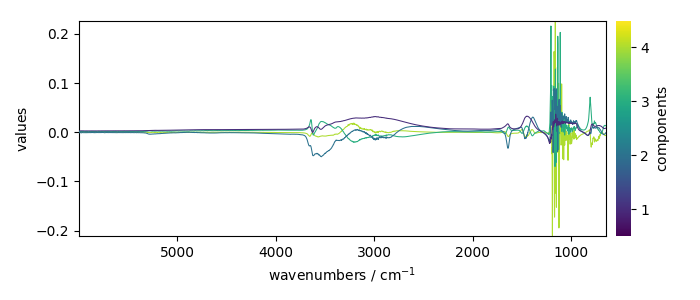
Displays 4 loadings
pca.loadings[:4].plot(legend=True)
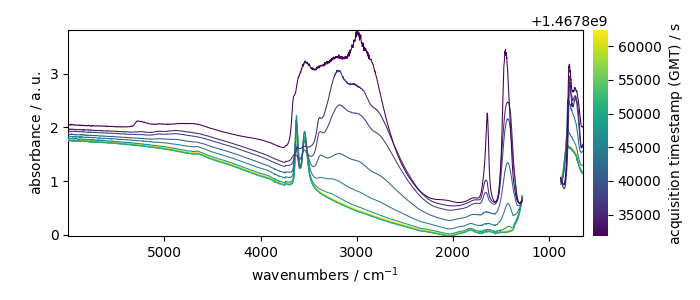
Here we do a masking of the saturated region between 882 and 1280 cm^-1
dataset[
:, 882.0:1280.0
] = scp.MASKED # remember: use float numbers for slicing (not integer)
dataset.plot()
Apply the PCA model
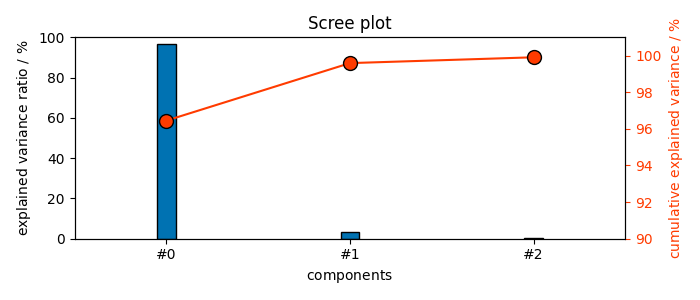
pca = scp.PCA(n_components=0.999)
pca.fit(dataset)
pca.n_components
3
As seen above, now only 4 components instead of 23 are necessary to 99.9% of explained variance.
(<Matplotlib Axes object>, <Axes: xlabel='components $\\mathrm{}$', ylabel='cumulative explained variance $\\mathrm{/\\ \\mathrm{\\%}}$'>)
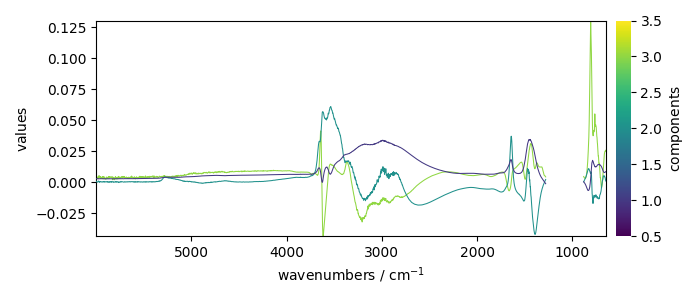
Displays the loadings
pca.loadings.plot(legend=True)
Let’s plot the scores
scores = pca.transform()
pca.scoreplot(scores, 1, 2)
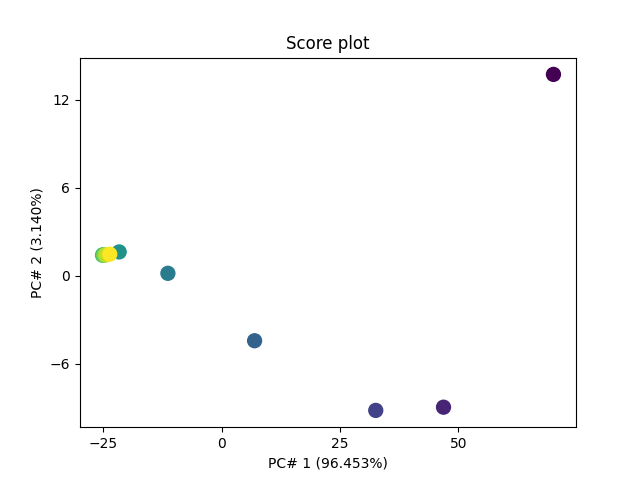
<Axes: title={'center': 'Score plot'}, xlabel='PC# 1 (96.453%)', ylabel='PC# 2 (3.140%)'>
Labeling scoreplot with spectra labels Our dataset has already two columns of labels for the spectra but there are little too long for display on plots.
array([[ 2016-07-06 19:03:14+00:00, vz0466.spa, Wed Jul 06 21:00:38 2016 (GMT+02:00)],
[ 2016-07-06 19:53:14+00:00, vz0471.spa, Wed Jul 06 21:50:37 2016 (GMT+02:00)],
...,
[ 2016-07-07 02:43:15+00:00, vz0512.spa, Thu Jul 07 04:40:39 2016 (GMT+02:00)],
[ 2016-07-07 03:33:17+00:00, vz0517.spa, Thu Jul 07 05:30:41 2016 (GMT+02:00)]], shape=(11, 2), dtype=object)
So we define some short labels for each component, and add them as a third column:
labels = [lab[:6] for lab in dataset.y.labels[:, 1]]
scores.y.labels = labels # Note this does not replace previous labels,
# but adds a column.
now display thse
pca.scoreplot(scores, 1, 2, show_labels=True, labels_column=2)
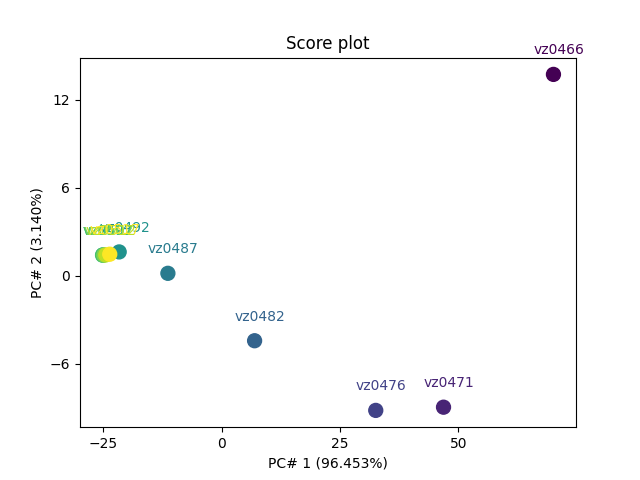
<Axes: title={'center': 'Score plot'}, xlabel='PC# 1 (96.453%)', ylabel='PC# 2 (3.140%)'>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: (0 minutes 1.507 seconds)