Note
Go to the end to download the full example code.
Loading Bruker OPUS files
Here we load an experimental Bruker OPUS files and plot it.
import spectrochempy as scp
Z = scp.read_opus(
["test.0000", "test.0001", "test.0002", "test.0003"], directory="irdata/OPUS"
)
Z
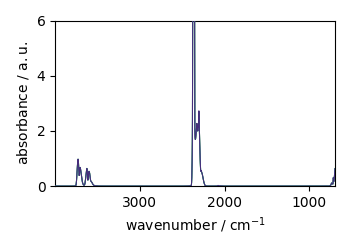
plot it
Z.plot()
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
scp.show()
Total running time of the script: (0 minutes 0.204 seconds)