Note
Go to the end to download the full example code.
Reading SPC format files
This example shows reading of ‘Galactic Industries’ SPC data file format. (.spc format).
First we need to import the spectrochempy API package
import spectrochempy as scp
The SPC format is a proprietary format from Galactic Industries. It is used for storing spectroscopic data. The SPC format is a binary file format that can store multiple spectra in a single file. The SPC format is widely used in the field of spectroscopy and is supported by many spectroscopic software packages.
Reading single file
ex1 = scp.read_spc("galacticdata/BENZENE.SPC")
ex1.plot()
ex1
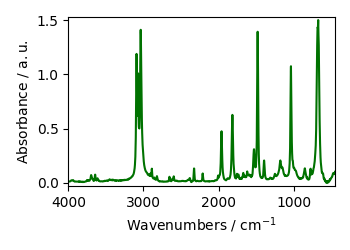
NDDataset: [float64] a.u. (shape: (y:1, x:1842))[/home/runner/.spectrochempy/testdata/galacticdata/BENZENE.SPC]
Summary
Memo: FT-IR Spectrum of Benzene, Capillary Film.
Data
Dimension `x`
Dimension `y`
reading SPC file with multiple subfiles with same x coordinates (they are merged by default)
ex2 = scp.read_spc("galacticdata/CONTOUR.SPC")
ex2.plot()
ex2
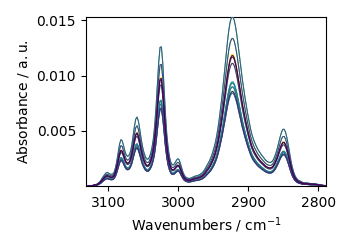
NDDataset: [float64] a.u. (shape: (y:19, x:179))[/home/runner/.spectrochempy/testdata/galacticdata/CONTOUR.SPC]
Summary
Memo: Subfiles are the Same Portion of POLYS Multiplied by a Sine Wave.
Data
[5.217e-05 4.488e-05 ... 6.45e-05 5.217e-05]
...
[5.243e-05 4.51e-05 ... 6.482e-05 5.243e-05]
[5.335e-05 4.589e-05 ... 6.596e-05 5.335e-05]] a.u.
Dimension `x`
Dimension `y`
Reading SPC file with multiple subfiles with different x coordinates (they are not merged)
ex3 = scp.read_spc("galacticdata/DRUG_SAMPLE_PEAKS.SPC")
for nd in ex3:
nd.plot_bar(width=0.1, clear=False)
ex3
# This ends the example ! The following line can be uncommented if no plot shows when
# running the .py script with python
# scp.show()
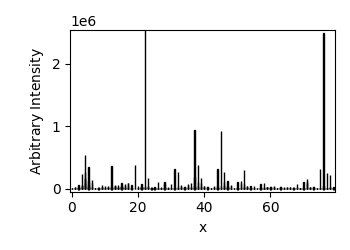
List (len=6, type=NDDataset)
0: NDDataset: [float64] unitless (shape: (y:1, x:124))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
1: NDDataset: [float64] unitless (shape: (y:1, x:141))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
2: NDDataset: [float64] unitless (shape: (y:1, x:237))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
3: NDDataset: [float64] unitless (shape: (y:1, x:237))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
4: NDDataset: [float64] unitless (shape: (y:1, x:253))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
5: NDDataset: [float64] unitless (shape: (y:1, x:80))[/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC]
Summary
Memo: Multifile of Peak Mass Spectra from Drug Sample.spcFormat
Log Text:
---------
[FILEINFO]
1999/10/14 14:10 W = Modified Date/Time (FILEINFO.AB) User=brigitte
Version = 5.22
[END FILEINFO]
[GC-MS]
Version = 1.10
2000/09/12 12:44 I = Interpolated (GCMS.AB) User=Brigitte
2000/09/12 12:44 T = Truncated (GCMS.AB) User=Brigitte
Original File = drug sample.spc
Operation: Save TIC peaks to Multifile
Peak Spectra:
Subfile # Left Bsln Center Z Right Bsln
74 4.66900015 4.75843334 4.85790014
176 5.68198347 5.77180004 5.81143332
187 5.82154989 5.88114977 5.98111677
242 6.33855009 6.42843342 6.52845001
279 6.70749998 6.79773331 6.89788342
306 6.9770999 7.06698322 7.16693354
[END GC-MS]
[FILEINFO]
2000/09/12 12:45 W = Modified Date/Time (FILEINFO.AB) User=Brigitte
Version = 6.00
[END FILEINFO]
/home/runner/.spectrochempy/testdata/galacticdata/DRUG_SAMPLE_PEAKS.SPC.
Data
Total running time of the script: (0 minutes 1.080 seconds)