Note
Go to the end to download the full example code.
Processing Relaxation measurement
Processing NMR spectra taken for relaxation measurements
Import API
import spectrochempy as scp
# short version of the unit registry
U = scp.ur
Importing a pseudo 2D NMR spectra
Define the folder where are the spectra
datadir = scp.preferences.datadir
nmrdir = datadir / "nmrdata" / "bruker" / "tests" / "nmr"
dataset = scp.read_topspin(nmrdir / "relax" / "100" / "ser", use_list="vdlist")
Analysing the data
Print dataset summary
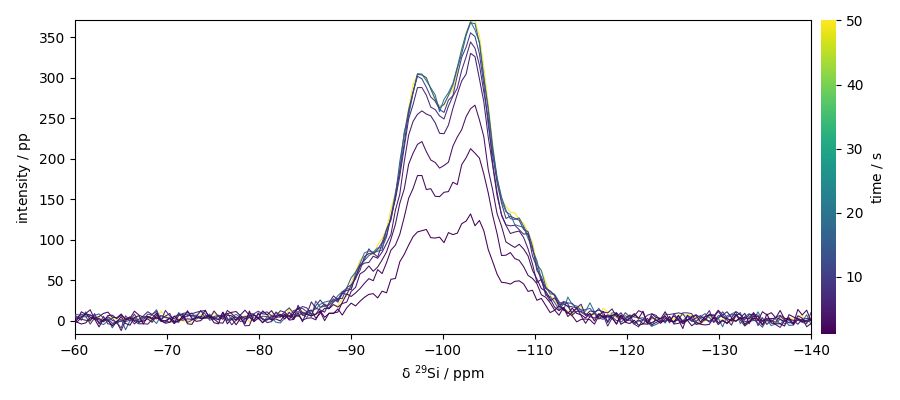
Plot the dataset
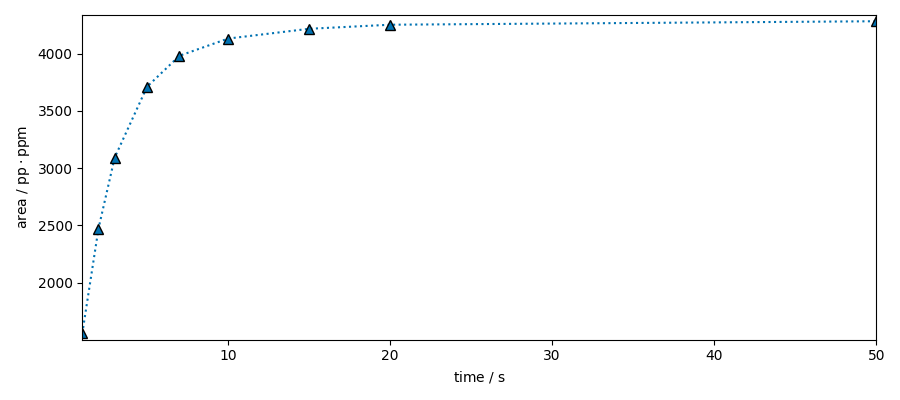
Integrate a region
dsint = ds[:, -90.0:-115.0].simpson()
dsint.plot(marker="^", ls=":")
dsint.real
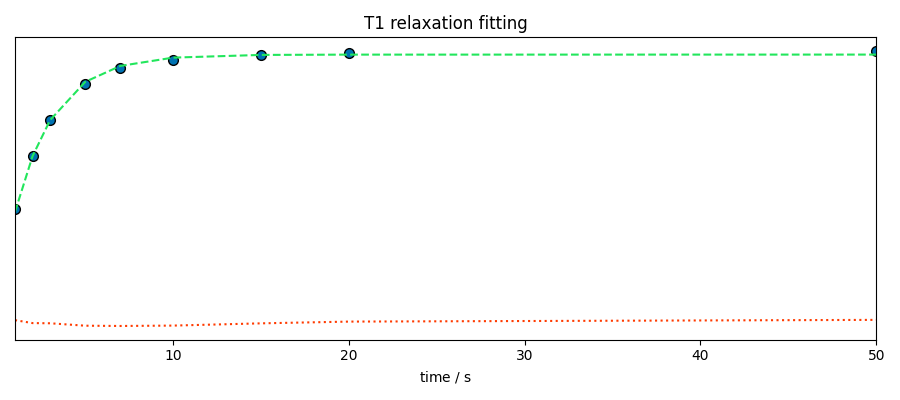
Fit a model
create an Optimize object using a simple leastsq method
fitter = scp.Optimize(log_level="INFO", method="leastsq")
Define the model to fit
def T1_model(t, I0, T1): # no underscore in parameters names.
# T1 relaxation model
import numpy as np
I = I0 * (1 - np.exp(-t / T1))
return I
Add the model to the fitter usermodels as it it not a built-in model
fitter.usermodels = {"T1_model": T1_model}
Define the parameter variables using a script (parameter: value, low_bound, high_bound) no underscore in parameters names.
fitter.script = """
MODEL: T1
shape: T1_model
$ I0: 1000.0, 1, none
$ T1: 2.0, 0.1, none
"""
Performs the fit
<spectrochempy.analysis.curvefitting.optimize.Optimize object at 0x7f0736814190>
som = fitter.predict()
som
fitter.plotmerit(dsint, som, method="scatter", title="T1 relaxation fitting")
This ends the example ! The following line can be removed or commented
when the example is run as a notebook (ipynb).
# scp.show()
Total running time of the script: (0 minutes 0.578 seconds)