Note
Go to the end to download the full example code.
NDDataset baseline correction
In this example, we perform a baseline correction of a 2D NDDataset
interactively, using the multivariate method and a pchip/polynomial interpolation.
For comparison, we also use the asls`and `snip models.
As usual we start by importing the useful library, and at least the spectrochempy library.
import spectrochempy as scp
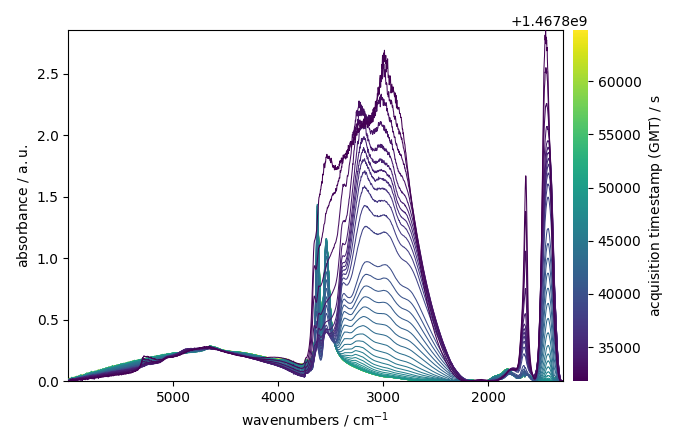
Load data
datadir = scp.preferences.datadir
nd = scp.read_omnic(datadir / "irdata" / "nh4y-activation.spg")
Do some slicing to keep only the interesting region
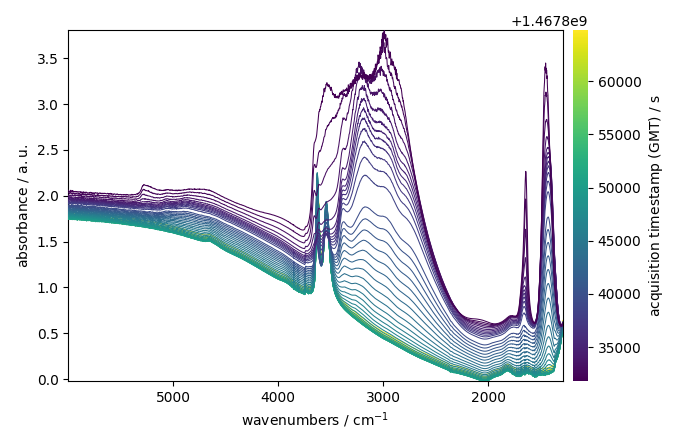
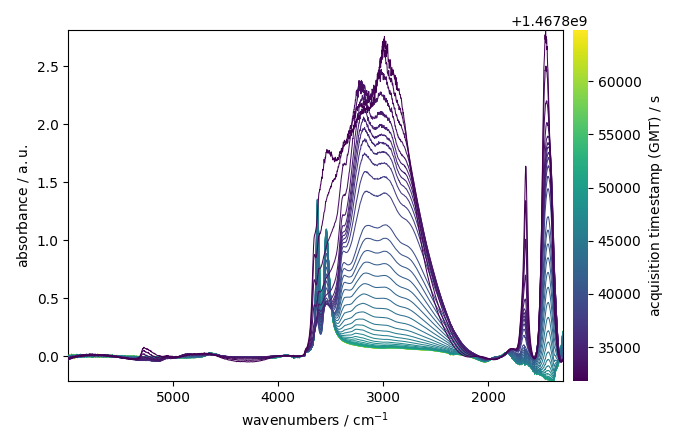
Plot the dataset
ndp.plot()
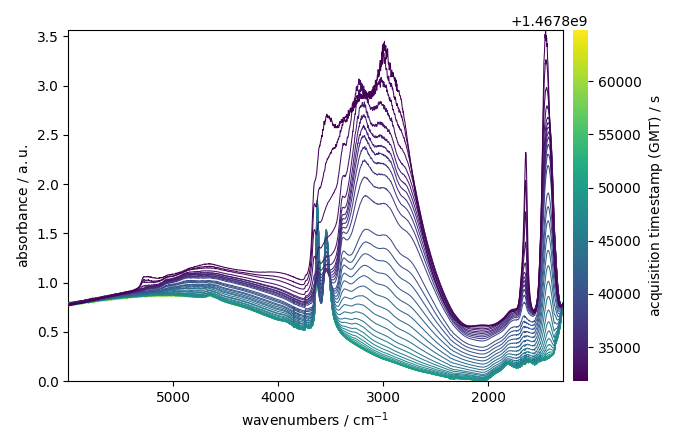
Remove a basic linear baseline using basc:
Make it positive
Define the Baseline object for a multivariate baseline correction model.
The n_components parameter is the number of components to use for the
multivariate baseline correction. The model parameter is the baseline
correction model to use, here a pchip interpolation (piecewise cubic
Hermite interpolation).
blc = scp.Baseline(
log_level="WARNING",
multivariate=True, # use a multivariate baseline correction approach
model="polynomial", # use a polynomial model
order="pchip", # with a pchip interpolation method
n_components=5,
)
Now we select the regions ( ranges ) to use for the baseline correction.
blc.ranges = [
[1556.30, 1568.26],
[1795.00, 1956.75],
[3766.03, 3915.81],
[4574.26, 4616.04],
[4980.10, 4998.01],
[5437.52, 5994.70],
]
We can now fit the baseline correction model to the data:
<spectrochempy.processing.baselineprocessing.baselineprocessing.Baseline object at 0x7f853aacdf90>
The baseline is now stored in the baseline attribute of the processor:
(note that the baseline is a NDDataset too).
The corrected dataset (the dataset after the baseline subtraction) is
stored in the corrected attribute of the processor:
Plot the result of the correction
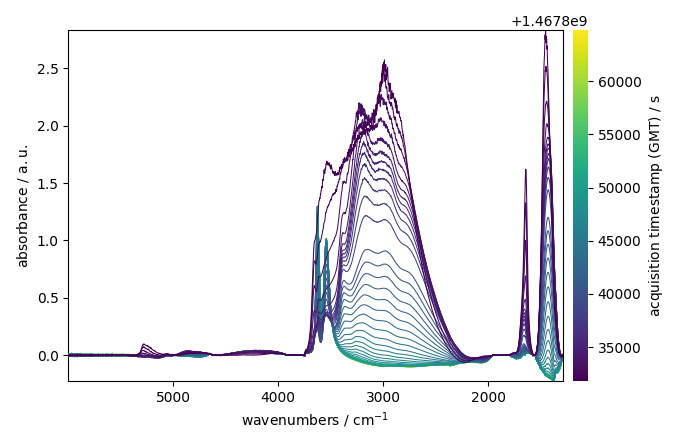
We can have a more detailed representation using plot
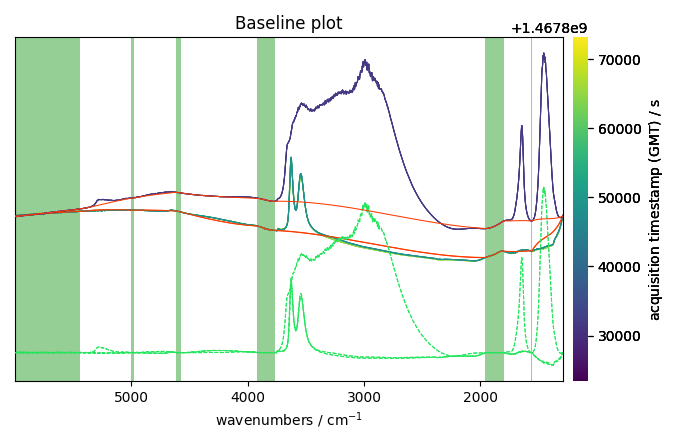
We can also plot the baseline and the corrected dataset together: for some individual spectra to, for example, check the quality of the correction:
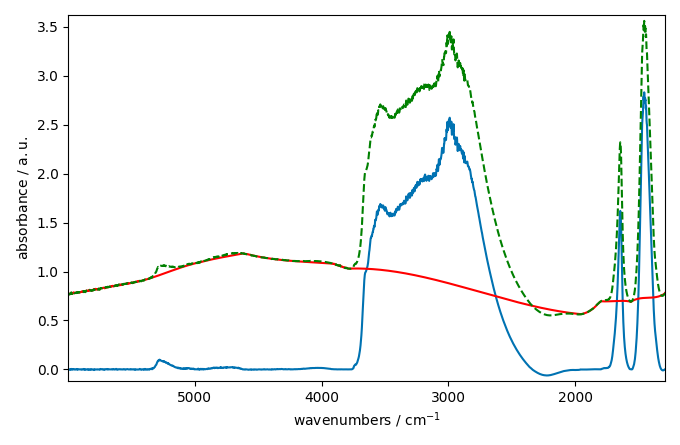
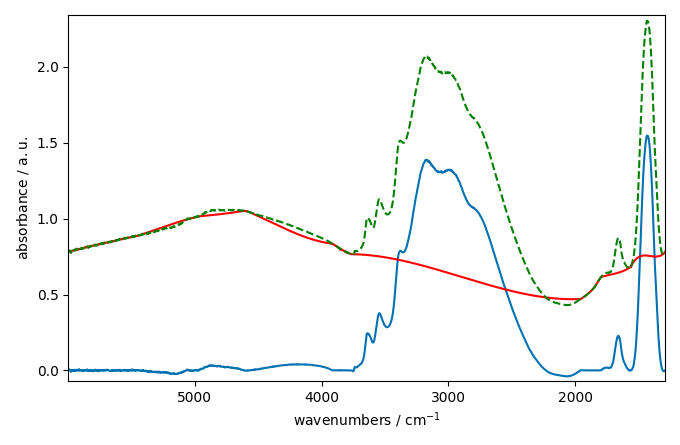
The baseline correction looks ok in some part of the spectra
but not in others where the variation seems a little to rigid.
This is may be due to the fact that the pchip interpolation
is perhaps not the best choice for this dataset. We can try to use a
n-th degree polynomial model instead:
We don’t need to redefine a new Baseline object, we can just change the model and the order of the polynomial:
and fit again the baseline correction model to the data:
blc.fit(ndp)
baseline = blc.baseline
corrected = blc.corrected
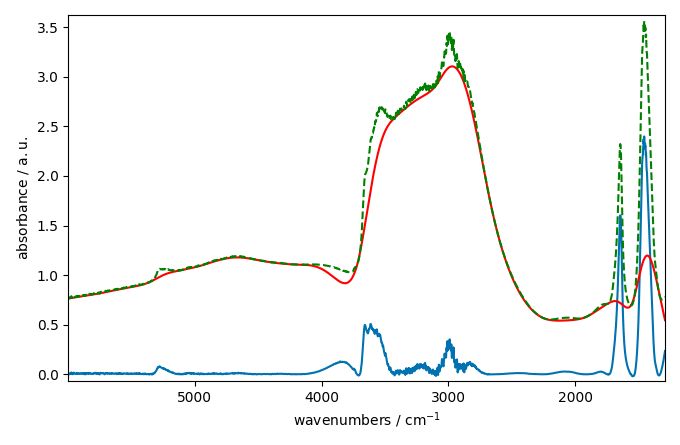
corrected[0].plot()
baseline[0].plot(clear=False, color="red", ls="-")
ndp[0].plot(clear=False, color="green", ls="--")
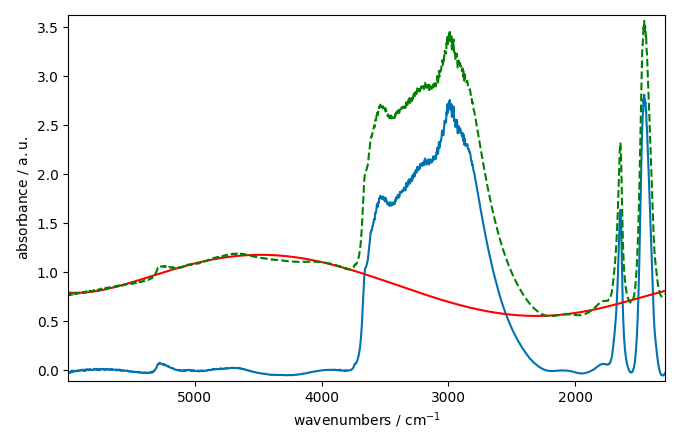
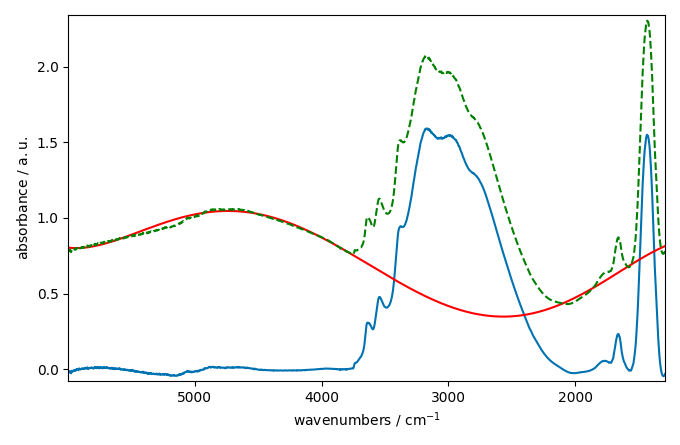
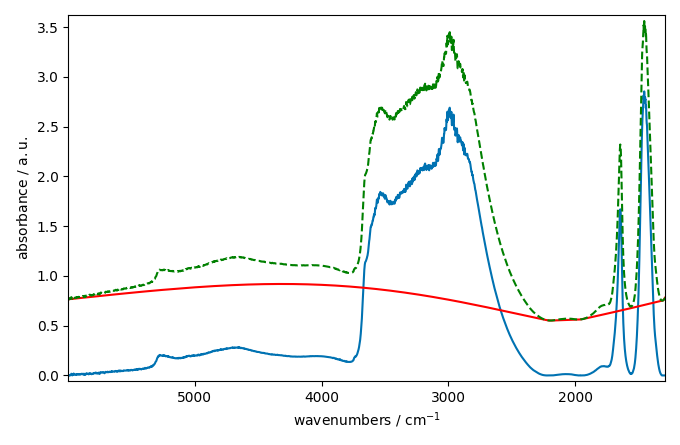
This looks better and smoother. But not perfect.
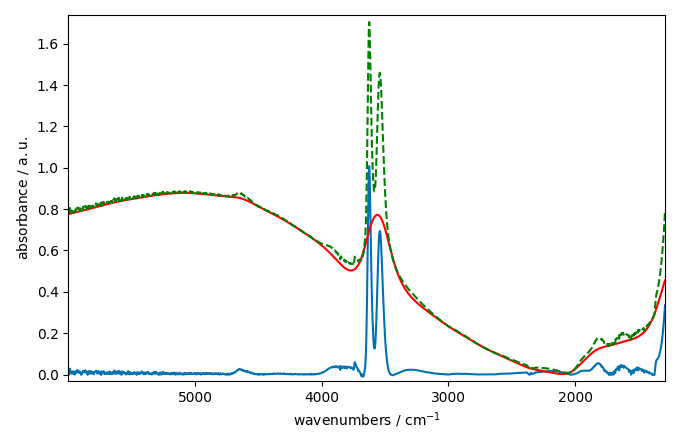
We can also try to use a asls (Asymmetric Least Squares) model
instead. This model is based on the work of Eilers and Boelens (2005)
and performs a baseline correction by iteratively fitting asymmetrically
weighted least squares regression curves to the data.
The asls model has two parameters: mu and assymetry.
The mu parameter is a regularisation parameters which control
the smoothness of the baseline. The larger mu is, the smoother
the baseline will be. The assymetry parameter is a parameter
which control the assymetry if the AsLS algorithm.
blc.multivariate = False # use a sequential approach
blc.model = "asls"
blc.mu = 10**9
blc.asymmetry = 0.002
blc.fit(ndp)
/home/runner/work/spectrochempy/spectrochempy/src/spectrochempy/processing/baselineprocessing/baselineprocessing.py:528: SparseEfficiencyWarning: spsolve requires A be CSC or CSR matrix format
z = spsolve(C, w * y)
<spectrochempy.processing.baselineprocessing.baselineprocessing.Baseline object at 0x7f853aacdf90>
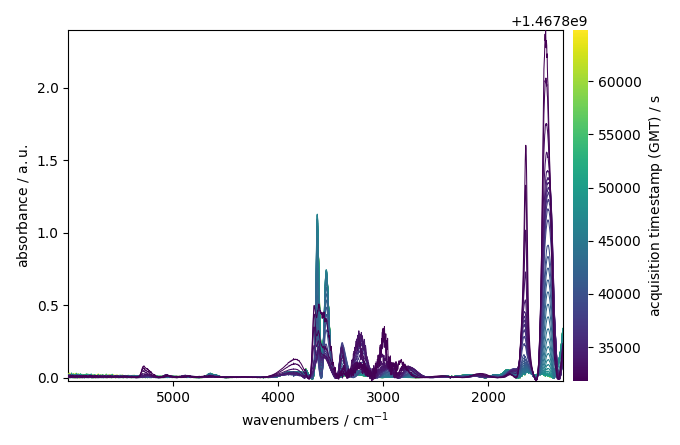
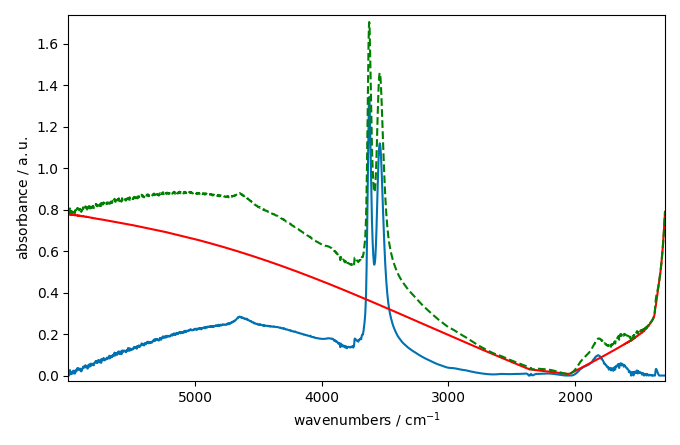
Finally, we will use the snip model
blc.multivariate = False # use a sequential approach
blc.model = "snip"
blc.snip_width = 200
blc.fit(ndp)
<spectrochempy.processing.baselineprocessing.baselineprocessing.Baseline object at 0x7f853aacdf90>
This ends the example ! The following line can be uncommented if no plot shows when running the .py script with python
# scp.show()
Total running time of the script: (0 minutes 5.822 seconds)